Nucleotide Sequence Databases
Understanding RefSeq
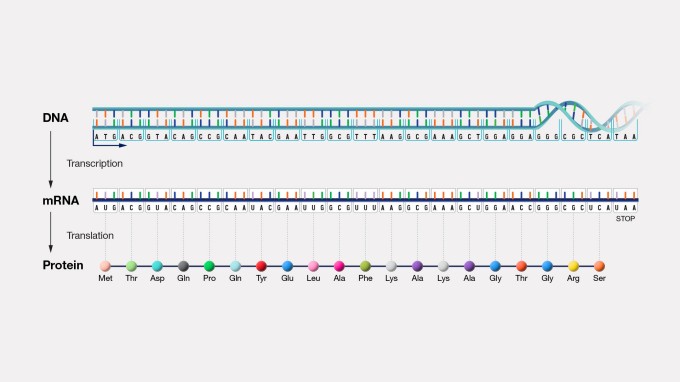
Image: NHGRI
Learning objectives:
- Gaining basic understading of RefSeq Database.
Navigating the Complexity of DNA Sequence Databases: A Guide to NCBI's RefSeq - A Database of Reference Sequences
In the rapidly evolving field of genomics, the production of DNA sequence data is occurring at an unprecedented pace, resulting in considerable redundancy within major sequence databases. This redundancy challenges researchers, making it difficult to discern the most accurate and reliable sequence information. To address this issue, the National Center for Biotechnology Information (NCBI) has developed RefSeq, a comprehensive project that provides a single reference sequence for each component of the central dogma - DNA to RNA to protein.
The Reference Sequence (RefSeq) database, an open-access initiative established in 2000 by the National Center for Biotechnology Information (NCBI), serves as a meticulously annotated and curated collection of nucleotide sequences (DNA, RNA) and their associated protein products obtained from the INSDC databases (GenBank, the European Nucleotide Archive, ENA, and the DNA Data Bank of Japan, DDBJ). Unlike GenBank, RefSeq provides a singular record for each natural biological molecule (DNA, RNA, or protein) within major organisms, from viruses and bacteria to eukaryotes, focusing on significant organisms with substantial data, currently encompassing almost 144,000 distinct "named" organisms as of November 2023.
Understanding the Challenge. The wealth of sequence data from systematic sequencing projects and individual laboratories brings specific challenges. A single biological entity may be represented by multiple entries across various databases, leading to confusion for end users. Additionally, distinguishing between experimentally determined sequences and computational predictions can be complicated.
RefSeq: A Non-Redundant Solution. NCBI's RefSeq project stands out as a pioneering initiative to overcome the challenges posed by redundant sequence data. The primary goal of RefSeq is to offer a non-redundant reference sequence for each molecule in the central dogma, encompassing DNA, mRNA, and protein. The uniqueness of RefSeq extends beyond its non-redundant nature; each entry includes comprehensive biological attributes of the gene, gene transcript, or protein.
Manually Curated Sequences
RefSeq is a precious and indispensable resource in genomics due to its meticulous manual curation of nucleotide sequences (DNA, RNA) and their associated protein products. RefSeq employs a rigorous curation process that combines experimental evidence, computational predictions, and manual curation. This multi-faceted approach ensures the accuracy and reliability of the sequences contained in the database. By providing a curated collection, RefSeq minimizes the risk of errors and inconsistencies often associated with uncurated or automated datasets.
The curated sequences in RefSeq help reduce ambiguity and confusion associated with poorly annotated or conflicting data. The transparent and standardized representation of genomic information minimizes misinterpretations, supporting more robust scientific conclusions.
The curated annotations include information about exons, introns, coding sequences, untranslated regions (UTRs), and other critical genomic features. This depth of knowledge is invaluable for understanding the functional elements of genes.
RefSeq also integrates genomic, transcriptomic, and proteomic information into a cohesive dataset. This integration allows researchers to systematically explore the relationships between genes, transcripts, and proteins. The hierarchical structure of RefSeq's notation format enables users to navigate through different levels of genomic information seamlessly.
The clinical domain. The manually curated sequences in RefSeq hold particular significance in the clinical domain. Researchers and clinicians rely on accurate and curated genomic information for applications such as disease diagnosis, identification of genetic variations, and understanding the molecular basis of disorders. The reliability of RefSeq is crucial in clinical genomics, where precision and accuracy are paramount.
Overall, the curated nature of RefSeq promotes standardization in genomic annotations. Researchers and laboratories worldwide can rely on RefSeq as a consistent reference, facilitating standardized analyses across different studies and experiments. This standardization is crucial for comparing and integrating genomic information from diverse sources.
Key Features of RefSeq
Linkage of Sequences: Nucleotide and protein sequences in RefSeq are explicitly linked, providing a holistic view of the molecular information.
Ongoing Curation: RefSeq entries undergo continuous curation, guaranteeing that the information remains up-to-date with the latest advancements in genomics.
Taxonomic Range: RefSeq entries cover a broad taxonomic range, reflecting the diversity of biological entities and ensuring relevance across different species.
RefSeq Accession Numbers
RefSeq entries are distinguishable from other databases, such as GenBank, through a unique accession number series. The format follows a "2 + 6" structure, where a two-letter code indicates the type of reference sequence, followed by an underscore and a six-digit number. Experimentally determined sequence data are denoted as NT (Genomic contigs), NM (mRNAs), and NP (Proteins). At the same time, those derived from genome annotation efforts are marked as XM (Model mRNAs) and XP (Model proteins).
Differentiating "N" and "X" Numbers. Understanding the distinction between "N" numbers and "X" is crucial. "N" numbers represent experimentally determined sequences, providing higher confidence in their accuracy. On the other hand, "X" numbers indicate computational predictions derived from raw DNA sequences, requiring a more cautious interpretation.
Notable categories include:
| AC | Complete genomic molecule (alternate assembly) |
| AP | Annotated on AC_alternate assembly |
| YP | Annotated on genomic molecules without an instantiated transcript record |
| NC | Complete genomic molecules |
| NG | Incomplete genomic regions |
| NZ | Complete genomes and unfinished WGS data |
| NT | Contig or scaffold (clone based or WGS) | NM | mRNA |
| NR | ncRNA |
| NP | Protein |
| NW | Contig or scaffold (primarily WGS) |
| XM | Predicted mRNA model |
| XR | Predicted ncRNA model |
| XP | Predicted Protein model (eukaryotic sequences) |
| WP | Predicted Protein model (prokaryotic sequences) |
The curation status of RefSeq Records
The RefSeq records have varied curation status levels. You can find the curation status of RefSeq entries in the COMMENT area of the record.
In the RefSeq database, the reliability of information is indicated by different status categories assigned to each record. The status categories can be broadly classified into two sets: one represents records that have undergone some level of review or validation, and the other includes records that are predictions or have yet to be individually reviewed.
More reliable status categories (have undergone some level of review or validation):
1. REVIEWED: This status indicates that NCBI staff or collaborators have reviewed the RefSeq record. The review process involves assessing available sequence data and relevant literature. These records are more reliable as they have undergone a quality check.
2. VALIDATED: This status indicates that the RefSeq record has been reviewed to establish the preferred sequence standard. Although validated, these records may be subject to final review for additional functional information.
Less reliable status categories (predictions or not yet individually reviewed):
1. MODEL: These records are generated by the NCBI Genome Annotation pipeline and are not subject to individual review or revision between annotation runs. They are based on computational predictions.
2. INFERRED: This status indicates that genome sequence analysis has predicted the RefSeq record but lacks experimental evidence. Homology data may partially support it.
3. PREDICTED: Records in this category have yet to be subject to individual review, and some aspects of the record are predicted. They are based on computational methods without thorough manual validation.
4. PROVISIONAL: RefSeq records in this category have yet to be subject to individual review. The initial sequence-to-gene association has been established by outside collaborators or NCBI staff, indicating a lower reliability level than reviewed records.
WGS (Whole Genome Shotgun) records represent a sequence not individually reviewed or revised between updates. They may need to be more reliable in terms of detailed annotation.
Observing these differences is important because it helps you gauge the confidence level in the accuracy and completeness of the data. Reviewed and validated records are generally more trustworthy, while predicted or provisional records should be interpreted cautiously and may require further experimental validation. You may find the RefSeq status codes in the COMMENT section of a record (Figure 1.).
| Status Code | Description |
|---|---|
| MODEL | The RefSeq record is provided by the NCBI Genome Annotation pipeline and is not subject to individual review or revision between annotation runs. |
| INFERRED | The RefSeq record has been predicted by genome sequence analysis, but it is not yet supported by experimental evidence. The record may be partially supported by homology data. |
| PREDICTED | The RefSeq record has not yet been subject to individual review, and some aspect of the RefSeq record is predicted. |
| PROVISIONAL | The RefSeq record has not yet been subject to individual review. The initial sequence-to-gene association has been established by outside collaborators or NCBI staff. |
| REVIEWED | The RefSeq record has been reviewed by NCBI staff or by a collaborator. The NCBI review process includes assessing available sequence data and the literature. Some RefSeq records may incorporate expanded sequence and annotation information. |
| VALIDATED | The RefSeq record has undergone an initial review to provide the preferred sequence standard. The record has not yet been subject to final review at which time additional functional information may be provided. |
| WGS | The RefSeq record is provided to represent a collection of whole genome shotgun sequences. These records are not subject to individual review or revisions between genome updates. |
See more in The NCBI online Handbook, Chapter 18 The Reference Sequence (RefSeq) Database.
LOCUS NM_001007083 1099 bp mRNA linear VRT 16-DEC-2021
DEFINITION Gallus gallus interleukin 3 (IL3), mRNA.
ACCESSION NM_001007083
VERSION NM_001007083.2
KEYWORDS RefSeq.
SOURCE Gallus gallus (chicken)
ORGANISM Gallus gallus
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda;
Coelurosauria; Aves; Neognathae; Galloanserae; Galliformes;
Phasianidae; Phasianinae; Gallus.
REFERENCE 1 (bases 1 to 1099)
AUTHORS Avery S, Rothwell L, Degen WD, Schijns VE, Young J, Kaufman J and
Kaiser P.
TITLE Characterization of the first nonmammalian T2 cytokine gene
cluster: the cluster contains functional single-copy genes for
IL-3, IL-4, IL-13, and GM-CSF, a gene for IL-5 that appears to be a
pseudogene, and a gene encoding another cytokinelike transcript,
KK34
JOURNAL J Interferon Cytokine Res 24 (10), 600-610 (2004)
PUBMED 15626157
REMARK GeneRIF: the chicken genome encodes genes for the homologs of
mammalian interleukin-3 (IL-3), IL-4, IL-5, IL-13, and
granulocyte-macrophage colony-stimulating factor (GM-CSF)
COMMENT VALIDATED REFSEQ: This record has undergone validation or
preliminary review. The reference sequence was derived from
JAENSK010000256.1.
...
Figure 1. RefSeq flatfile header where the COMMENT line has the status code 'VALIDATED.'
RefSeq Flatfiles and FASTA Files
Example of RefSeq flatfile - same format as Genbank flatfiles. Note that the sequence and annotations is not shown on this file due to its larges size. You can see the entire file online at NCBI and you can use the "Customize view" panel to change the display on the left side panel to display the complete file.
LOCUS NC_000067 195154279 bp DNA linear CON 10-APR-2023
DEFINITION Mus musculus strain C57BL/6J chromosome 1, GRCm39.
ACCESSION NC_000067
VERSION NC_000067.7
DBLINK BioProject: PRJNA169
Assembly: GCF_000001635.27
KEYWORDS RefSeq.
SOURCE Mus musculus (house mouse)
ORGANISM Mus musculus
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Glires; Rodentia; Myomorpha;
Muroidea; Muridae; Murinae; Mus; Mus.
REFERENCE 1 (bases 1 to 195154279)
AUTHORS Church,D.M., Schneider,V.A., Graves,T., Auger,K., Cunningham,F.,
Bouk,N., Chen,H.C., Agarwala,R., McLaren,W.M., Ritchie,G.R.,
Albracht,D., Kremitzki,M., Rock,S., Kotkiewicz,H., Kremitzki,C.,
Wollam,A., Trani,L., Fulton,L., Fulton,R., Matthews,L.,
Whitehead,S., Chow,W., Torrance,J., Dunn,M., Harden,G.,
Threadgold,G., Wood,J., Collins,J., Heath,P., Griffiths,G.,
Pelan,S., Grafham,D., Eichler,E.E., Weinstock,G., Mardis,E.R.,
Wilson,R.K., Howe,K., Flicek,P. and Hubbard,T.
TITLE Modernizing reference genome assemblies
JOURNAL PLoS Biol. 9 (7), e1001091 (2011)
PUBMED 21750661
REFERENCE 2 (bases 1 to 195154279)
AUTHORS Church,D.M., Goodstadt,L., Hillier,L.W., Zody,M.C., Goldstein,S.,
She,X., Bult,C.J., Agarwala,R., Cherry,J.L., DiCuccio,M.,
Hlavina,W., Kapustin,Y., Meric,P., Maglott,D., Birtle,Z.,
Marques,A.C., Graves,T., Zhou,S., Teague,B., Potamousis,K.,
Churas,C., Place,M., Herschleb,J., Runnheim,R., Forrest,D.,
Amos-Landgraf,J., Schwartz,D.C., Cheng,Z., Lindblad-Toh,K.,
Eichler,E.E. and Ponting,C.P.
CONSRTM Mouse Genome Sequencing Consortium
TITLE Lineage-specific biology revealed by a finished genome assembly of
the mouse
JOURNAL PLoS Biol. 7 (5), e1000112 (2009)
PUBMED 19468303
REFERENCE 3 (bases 1 to 195154279)
CONSRTM Genome Reference Consortium
TITLE Direct Submission
JOURNAL Submitted (24-JUN-2020) NCBI, NIH, Bethesda, MD 20892, USA
COMMENT REFSEQ INFORMATION: The reference sequence is identical to
CM000994.3.
On Sep 22, 2020 this sequence version replaced NC_000067.6.
Assembly Name: GRCm39
The DNA sequence is composed of genomic sequence, primarily
finished clones that were sequenced as part of the Mouse Genome
Project. PCR products and WGS shotgun sequence have been added
where necessary to fill gaps or correct errors. All such additions
are manually curated by GRC staff. For more information see:
https://genomereference.org.
##Genome-Annotation-Data-START##
Annotation Provider :: NCBI RefSeq
Annotation Status :: Updated annotation
Annotation Name :: GCF_000001635.27-RS_2023_04
Annotation Pipeline :: NCBI eukaryotic genome annotation
pipeline
Annotation Software Version :: 10.1
Annotation Method :: Best-placed RefSeq; Gnomon;
RefSeqFE; cmsearch; tRNAscan-SE
Features Annotated :: Gene; mRNA; CDS; ncRNA
Annotation Date :: 04/05/2023
##Genome-Annotation-Data-END##
FEATURES Location/Qualifiers
source 1..195154279
/organism="Mus musculus"
/mol_type="genomic DNA"
/strain="C57BL/6J"
/db_xref="taxon:10090"
/chromosome="1"
CONTIG join(gap(100000),gap(10000),gap(2890000),gap(50000),
NT_039170.9:1..82274824,gap(50000),NT_078297.8:1..109679455,
gap(100000))
//
FASTA-format
You can also obtain RefSeq sequences in FASTA-format. Here's an example, trucated due to the large size:
>NC_000067.7 Mus musculus strain C57BL/6J chromosome 1, GRCm39
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
...
TATCTTAATTAGTTTTTGAGTTCTCCAAAGCTATTTGCTCTCTGTGTTGTTAACCTGTACAAGACTGAAG
GTTCTTATTCCTATATCTTATTAATATTCACATTGACATTTTGATGTCTGCTTTCTATATTTTCCTAAAA
ATATTTTAAAGTACACACTATACAGACTTTTAATTTAATTCAGTTTTCTATTCAGGTAATATATTTTGAT
CACATTTACCCCTGCTTCAAAGTTGCTAGTATGAGATTATCCTAAATTTTTTATGAAGACACTTATTACT
ATGAACTCTCCTCCTAGTATTGATTTCATTGGGTCTCATAAGTTTGGGTATGTTGTGAATTTGTTTTCAT
...
RefSeqGene
RefSeqGene genes constitute a vital subset within the RefSeqs, designed as an integral component of the reference genome. This specialized subset comprises well-defined, complete genes meticulously curated as stable reference genes. Their primary purpose is establishing precise coordinates for all gene regions, encompassing promoters, introns, exons, and flanking regions. Additionally, RefSeqGene plays a pivotal role in delineating gene mutations and biologically significant variants, such as CNVs and SNPs, i.e., Copy Number Variations and Single Nucleotide Polymorphisms.
RefSeqGene, in contrast to the broader RefSeq database, distinguishes itself by providing gene-specific sequences for each recognized gene, presenting a comprehensive and complete set of regions associated with the gene structure. These sequences undergo meticulous alignment to reference chromosomes, ensuring their status as standard, normal alleles. RefSeqGene serves as a baseline reference, offering a robust foundation for gene sequences and facilitating precise analysis of genetic elements and variations in the context of the entire genome. This specialized resource is invaluable in research and clinical settings, contributing to a deeper understanding of genomic structures and their implications in health and disease.
References
-
O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O'Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016 Jan 4;44(D1):D733-45. doi: 10.1093/nar/gkv1189. PMID: 26553804; PMCID: PMC4702849.
-
Tatusova T, Ciufo S, Fedorov B, O'Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2014 Jan;42(Database issue):D553-9. doi: 10.1093/nar/gkt1274. PMID: 24316578; PMCID: PMC3965038.
-
Sayers EW, Bolton EE, Brister JR, Canese K, Chan J, Comeau DC, Farrell CM, Feldgarden M, Fine AM, Funk K, Hatcher E, Kannan S, Kelly C, Kim S, Klimke W, Landrum MJ, Lathrop S, Lu Z, Madden TL, Malheiro A, Marchler-Bauer A, Murphy TD, Phan L, Pujar S, Rangwala SH, Schneider VA, Tse T, Wang J, Ye J, Trawick BW, Pruitt KD, Sherry ST. Database resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023 Jan 6;51(D1):D29-D38. doi: 10.1093/nar/gkac1032. PMID: 36370100; PMCID: PMC9825438.
