Protein Sequence Databases
The NCBI Identical Protein Groups (IPG) Database

Learning objectives:
- Understand the Structure and Purpose of the IPG Database.
- Learn to Navigate the IPG Database.
- Interpret and Utilize IPG Data.
Introduction
The NCBI Identical Protein Groups (IPG) Database was created to simplify searching for protein information across vast and diverse datasets. Before its establishment, searching for a protein in the NCBI Protein database by name or symbol often led to multiple redundant records from different organisms, making it difficult to find specific protein information and hindering comparative and evolutionary studies.
To address this issue and help users find relevant protein data more efficiently, NCBI developed the IPG database. Its primary objective is to group identical proteins, reducing redundancy and simplifying the search and analysis. By doing so, researchers can focus on unique protein sequences and their characteristics without navigating through a long list of repetitive entries.
Each entry in the IPG database represents a unique protein translation found across various sources that NCBI encompasses, including multiple databases such as GenBank, RefSeq, INSDC, SwissProt, and PDB, ensuring a comprehensive and inclusive dataset. Integrating these sources into a single platform means that the IPG provides a complete view of protein sequences, offering insights that might be fragmented or overlooked when consulting individual databases.
The Structure of IPG
The IPG database is a collection of protein records derived from the NCBI's extensive protein sequence database. These records are systematically grouped based on their sequence identity. The primary aim is to cluster identical or highly similar proteins across different species, reflecting evolutionary relationships and functional similarities. This clustering is crucial for comparative genomics and evolutionary biology studies, as it allows scientists to trace the lineage and conservation of proteins across different organisms.
The IPG database organizes proteins hierarchically and relationally to efficiently manage and retrieve protein information. It sorts proteins into groups based on their sequence identity at the highest level. Each group further organizes proteins into subgroups, typically based on species or taxonomic classification. This hierarchical organization facilitates a nuanced analysis of proteins, enabling researchers to discern patterns and relationships that might be obscured in a less structured dataset.
The IPG database provides a wealth of information about each protein record, including the protein sequence, length, the source organism, and links to other databases and literature. Additionally, it links these records with other NCBI databases such as GenBank and PubMed, giving a detailed understanding of the protein's genetic context, functional annotations, and relevant scientific research.
The IPG database has many features and tools that help with bioinformatics research. Users can conduct complex searches to find proteins that meet specific criteria, use advanced algorithms to compare sequences and examine the evolutionary connections between proteins through phylogenetic trees and other graphical representations.
IPG Indexed Fields
The Identical Protein Groups (IPG) indexed fields, as outlined below (Table 1), serve as a comprehensive schema designed to categorize and facilitate the retrieval of protein-related data. See the Appendix for more detailed descriptions of the index fields.
| Description | Short Field | Description | Short Field |
|---|---|---|---|
| Accession (unique identifier) | ACCN | All Fields (cumulative) | ALL |
| Assembly (genomic data) | ASSM | Creation Date | CDAT |
| Division (classification) | DIV | Filter (data refinement) | FILT |
| BioProject link | GPRJ | Modification Date | MDAT |
| Molecular Weight | MLWT | Organism Count | OCNT |
| Organism | ORGN | Primary Accession | PACC |
| Properties | PROP | Protein Name | PROT |
| Protein Count | ProtCount | Sequence Length | SLEN |
| Title | TITL | Update Date | UDAT |
| Unique Identifier | UID | Text Word (searchable) | WORD |
Navigating the IPG Database
To begin a search, enter the specific name or symbol of the protein you want to find in the search bar at www.ncbi.nlm.nih.gov/ipg. For instance, you can search for "nADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1" [PROT]. This search will provide you with a wide range of information related to nADH proteins, as an example.
Navigating the vast landscape of protein data can be overwhelming, especially when initial searches yield a broad spectrum of results. Refining your search parameters becomes essential to address this. For instance, if you want to focus on the nADH specific to Mus musculus (mouse), you need a more targeted search phrase like "nADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1" [PROT] AND "Mus musculus" [ORGN]. Such specificity narrows down the results, making them more relevant and manageable (Figure 1).
Upon executing a search, various IPG records emerge, each representing a distinct protein. Selecting a record allows for an in-depth exploration of detailed protein information. The IPG Record Page is a critical component of this exploration, offering a summary that encompasses vital details like the protein's name, accession number, length, and taxonomic classification.
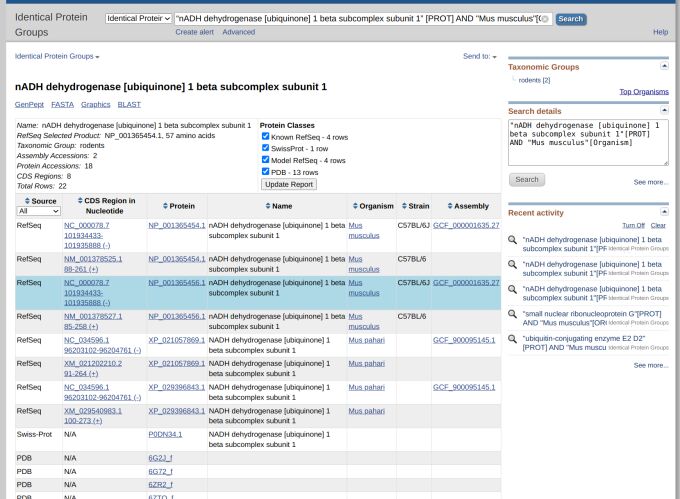
This page also features taxonomic groups that serve as navigational filters, leading you through the sequences of different members. Additional information such as source database, accession numbers, and organism associations for each protein further enriches this data.
The IPG database provides various tools and links that enhance research capabilities. The GenPept and FASTA formats offer alternative views of the protein sequence, facilitating visual and structural comprehension (See more about file formats in our tutorial The Data Format In Nucleotide Sequence Databases). The Sequence Viewer tool is particularly beneficial, allowing for a graphical representation of the protein RefSeq and associated features, which offers a deeper understanding of the protein's structure and function. The BLAST link is indispensable for comparative sequence analysis, enabling comparisons of your protein of interest against a comprehensive sequence database.
Effective search strategies in the IPG database involve specificity in search terms and using indexed fields like [PROT] for protein names or [ORGN] for organisms. This targeted approach ensures more refined and relevant search outcomes. A thorough exploration of the search results is also encouraged, as delving into the IPG records reveals extensive details about taxonomic groups and sequence specifics.
When creating advanced search queries in the IPG database, you can use indexed fields listed in Table 1 of the provided text. These fields include Accession (ACCN), Organism (ORGN), Protein Name (PROT), and many others, allowing you to refine your searches
Besides AND, you can also use Boolean operators OR or NOT. Note that the operators must be in uppercase.
Appendix
The fields indexed by Identical Protein Groups (IPG), as shown in Table 1, provide a complete schema to categorize and help retrieve protein-related data. These fields contain different attributes and metadata linked to database protein entries, allowing researchers to query and analyze the data efficiently. Each field has a shorthand notation, making it easier to reference in database queries or software applications.
- Accession (ACCN): This field holds the unique accession number assigned to each protein or protein group. It acts as a primary identifier that users can employ to fetch specific protein records.
- All Fields (ALL): This is a cumulative field that encompasses all other fields, providing a broad search parameter. Queries against this field will return results matching any of the indexed attributes.
- Assembly (ASSM): This field relates to the genomic assembly data from which the protein is derived. It links proteins to their corresponding genomic contexts, aiding in the study of gene-protein relationships.
- Creation Date (CDAT): The date the protein record was created. This field is crucial for tracking the newest entries or understanding the historical context of the data.
- Division (DIV): This categorizes proteins into different divisions or sections based on certain criteria, such as organism type or functional classification, facilitating more focused searches.
- Filter (FILT): A field used to apply specific filters to the data, potentially based on quality, relevance, or other criteria defined by the database or the user.
- BioProject (GPRJ): This links proteins to their respective BioProject, a collection or project within the NCBI database that has related biological data. It's useful for researchers looking at proteins within broader research contexts.
- Modification Date (MDAT): Similar to the creation date, this indicates the last date the protein record was modified, which is essential for understanding the currency and history of the data.
- Molecular Weight (MLWT): The calculated molecular weight of the protein. This physical characteristic is fundamental to many types of biochemical analysis.
- Organism Count (OCNT): Represents the number of different organisms in which the protein or protein group is found. This can indicate the ubiquity and evolutionary significance of the protein.
- Organism (ORGN): The scientific name of the organism from which the protein is derived. It's vital for studies focused on specific species or comparative analyses.
- Primary Accession (PACC): This might refer to the principal or original accession number assigned to the protein, distinct from secondary or alternative identifiers.
- Properties (PROP): A general field that can encompass various characteristics and annotations related to the protein, such as functional properties, structural classifications, or interaction data.
- Protein Name (PROT): The name or title given to the protein, often reflecting its function or characteristics. It's a key field for initial searches and identification.
- Protein Count (ProtCount): Indicates the number of proteins within a particular group or dataset. This can be useful in understanding the scope and scale of the data.
- Sequence Length (SLEN): The length of the amino acid sequence of the protein. It's a fundamental attribute that affects the protein's structure and function.
- Title (TITL): Often a descriptive title for the protein entry, which may provide quick insight into the protein's nature and role.
- Update Date (UDAT): Indicates when the record was last updated, helping users identify the most recent and potentially most accurate data.
- Unique Identifier (UID): A distinct number or code assigned to each protein record to ensure each entry is unique and retrievable.
- Text Word (WORD): This field is likely used for text-based searches, allowing users to query descriptive text or annotations associated with the protein records.
