Protein Sequence Databases
Introduction to Biological Databases
Learning objectives:
- Recognize Biological Data Complexity.
- Learn about the difficulties in organizing different types and sizes of biological data.
- Discover how unique numbers help track and manage changes in biological data over time.
- Understand how various databases work together to support bioinformatics research.
- Learn about how mistakes can spread in connected databases and how they try to keep data accurate.
- Get to know the difference between primary, secondary, and composite databases and what they do.
Introduction
Understanding the Complex Landscape of Biological Data in Bioinformatics
Have you ever wondered how scientists keep track of the immense and ever-growing amount of biological data? Biological data, with its intricate structures and relationships, demands sophisticated systems to ensure no critical detail is overlooked.
The Intricacy of Biological Data Representation
Consider the case of the National Center for Biotechnology Information (NCBI). It's not just handling simple strings of characters but representing biological sequences through complex systems, such as integer coordinate systems. Have you thought about what it means to attach different data types to each point in a sequence? The complexity mirrors the multifaceted nature of life itself, like the intricate arrangement of amino acids in proteins.
Challenges in Handling Biological Data
Imagine trying to categorize the vast diversity of life's building blocks. Biological data is not uniform; it varies significantly in size and the type of natural information it represents. This diversity requires a system and a flexible, adaptive approach to data management, accommodating the unique exceptions and irregularities found in biological structures.
Database Management and the Role of Accession Numbers
In this ever-evolving landscape, how do databases keep up? Databases maintain pace by utilizing unique identifiers called accession numbers to track and manage data.
GenBank, a prominent example, employs a system of accession and version numbers to meticulously track protein sequence evolutions, allowing researchers to access and compare historical data.
Real-World Scenario: Consider a research team submitting a novel protein sequence related to oncogenic pathways. Not only is this sequence archived, but as the scientific community studies it, they may contribute insights into its functional domains or evolutionary lineage.
Analogical Insight: Imagine a library where every new book edition is added alongside the old, continually building upon the existing knowledge. This is how these databases manage updates, adding new records with updated information without overwriting the old, preserving both the past and present data.
Similarly, the DNA Database of Japan (DDBJ) and the European Nucleotide Archive (ENA) use these identifiers to ensure every piece of data, from DNA sequences to nucleotide-level information, is traceable and accessible as it evolves. This approach supports collaborative research efforts across borders and ensures the reliability of genetic data.
Interconnectedness and the Big Picture
What does this all mean for the field of bioinformatics? It's a world of interconnected databases, each feeding into and building upon the other. Primary databases like GenBank, DDBJ, and ENA provide the raw data, which is refined and contextualized in secondary databases like InterPro, storing protein families, motifs, and domains. These databases are not isolated islands but part of a vast, interconnected ecosystem supporting various bioinformatics activities.
Discussion of Challenges and Solutions in Biological Data Management
Dealing with large and complex biological data presents several challenges for bioinformatics. The storage, retrieval, and analysis systems experience significant pressure due to the exponential growth of data. The data includes various hybrid data, such as genomic sequences and protein structures. The diversity of data formats also makes standardization a difficult task for researchers, who must confront inconsistency and incompatibility issues.
The pace at which knowledge and techniques evolve constantly increases, requiring frequent database updates, which can be expensive and time-consuming. Sharing sensitive medical data, personal genomes, and ancestry information is crucial to advancing medicine. However, strict privacy regulations, complex ownership rights, and inconsistent standards often hinder this, making global accessibility challenging. It is paramount to strike a mindful balance between open scientific inquiry and protecting personal privacy.
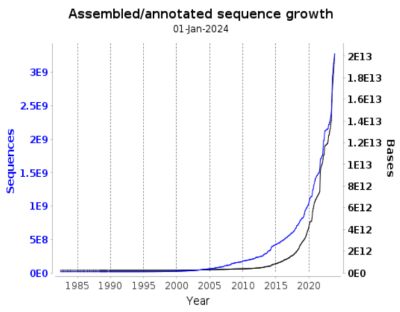
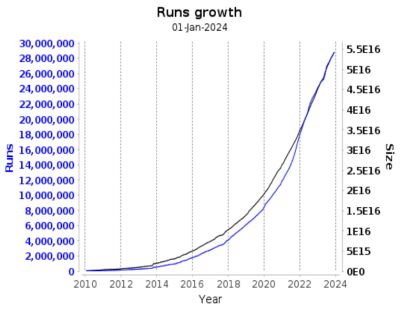
In response to challenges, advanced database systems are developed to handle growing data volume and complexity with robust, scalable, and flexible architectures.
Systems often use sophisticated indexing and querying mechanisms to enhance their efficiency. Ongoing efforts to standardize biological data formats are led by initiatives like the Human Genome Project, the Proteomics Standards Initiative (HUPO), and the International Nucleotide Sequence Database Collaboration (INSDC). These initiatives aim to ensure data consistency and interoperability.
Cloud computing has also transformed how we store and analyze data, rendering expensive infrastructure unnecessary. Additionally, artificial intelligence and machine learning techniques have become more prevalent in extracting valuable insights, predicting trends, and identifying new research areas in biological data management, representing a significant advancement in addressing the complexities of managing biological data.
Mitigating Error Propagation in Biological Databases: An Ongoing Challenge for Data Integrity
The interconnectedness of biological databases can lead to the spread of inaccuracies, a significant concern. For example, when one database contains an error in gene annotation, sequence identity, or functional data, it can be propagated through other databases that rely on it for updates.
While this interconnectedness benefits data accessibility and richness, it can also lead to misinformation. Even a single error can significantly impact research, diagnostics, and therapeutic strategies based on the data.
Databases are designed to tackle error propagation by implementing strict validation and cross-referencing mechanisms. However, the challenge persists due to the enormous size and complexity of the data. To tackle this issue, researchers and database curators must collaborate continuously to identify, correct, and prevent the spread of inaccuracies.
This requires a multifaceted approach, including algorithmic error detection, community reporting systems, and periodic review of database entries. Despite these efforts, the dynamic nature of biological data and ongoing discoveries mean that maintaining absolute accuracy is an ever-evolving challenge, highlighting the need for constant vigilance and improvement in data management practices.
Database Types in Bioinformatics: An Overview
In bioinformatics, understanding the diverse types of databases is crucial. We can broadly categorize these into three types: primary (archival), secondary (curated or knowledgebase), and composite (hybrid). Each has a specific role within Bioinformatics.
Primary Databases: The Archival Records
What They Are: Primary databases are the foundational layer of biological information. They store raw data from experimental outputs like DNA or protein sequences and structural information.
How They Work: This category is characterized by unprocessed, as-is data submissions, typically from researchers and laboratories worldwide. Each dataset is assigned a unique accession number upon submission, marking its official entry into the scientific record.
Examples: EMBL, GenBank, and DDBJ handle nucleotide sequences, ArrayExpress and GEO functional genomics data, and Protein Data Bank (PDB) stores coordinates of three-dimensional macromolecular structures.
Secondary Databases: The Curated Knowledgebases
What They Are: Secondary databases build upon the raw data in primary databases by adding value through curation, annotation, and integration.
Their Role includes processed and interpreted data, such as protein domain structures and pathway information. The curation process, often involving expert review, ensures the accuracy and relevance of the information.
Examples: UniProt Knowledgebase handles sequence and functional information on proteins, InterPro deals with protein families, motifs, and domains, and Ensembl aids research in comparative genomics, evolution, and regulation by annotating genes, aligning sequences, predicting regulatory functions, and compiling disease data.
Composite Databases: The Hybrid Systems
What They Are: Composite databases merge features from primary and secondary databases, offering a versatile resource for comprehensive data access.
How They're Used: They provide a user-friendly interface to query, retrieve, and analyze data from multiple sources, bridging the gap between raw and curated information, which is particularly useful for complex queries requiring cross-referencing between data types and sources.
Examples: The UCSC Genome Browser, developed by the University of California Santa Cruz, is a sophisticated visualization platform integrating genomic information from diverse sources.
Similarly, the National Center for Biotechnology Information's Entrez system is an exemplary composite database, offering a unified platform to query multiple databases, covering a spectrum of data types like nucleotide and protein sequences, protein structures, and scholarly articles.
Additionally, Expasy, an expansive portal curated by the Swiss Institute of Bioinformatics (SIB), grants access to over 160 databases and tools, supporting a wide array of research areas from genomics and proteomics to systems biology and medical chemistry, underscoring its role in the integration of bioinformatics resources.
Interconnectedness and the Big Picture
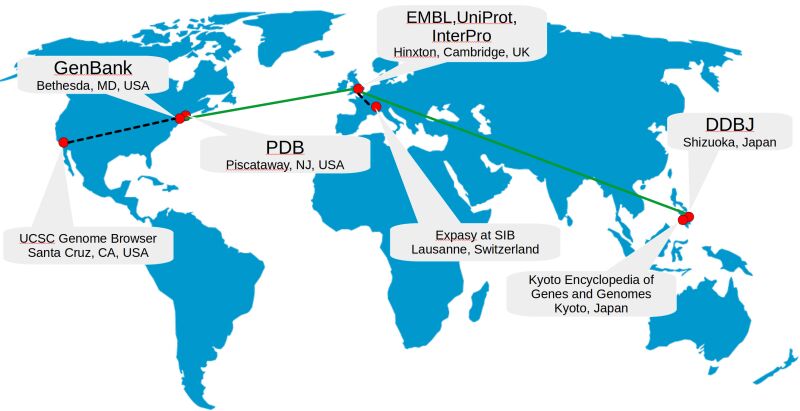
While each database type has its distinct role, its true power lies in its interconnectedness. Primary databases feed into secondary ones, which then integrate into composite databases for a multifaceted analysis, Thus creating a rich, layered ecosystem of information far more potent than the sum of its parts.
Data is stored, contextualized, and analyzed through this interconnected framework, providing a comprehensive platform for various Bioinformatics activities.
From genetic research and molecular modeling to epidemiological studies and drug discovery, the interconnected nature of these databases significantly amplifies their utility and impact. Therefore, Understanding each database type's unique contributions becomes crucial in leveraging their collective power effectively.
Primary databases are the repositories of raw data, the bedrock upon which further analysis is built. With their curated and annotated data, secondary databases provide the insights and contextual information that transform raw data into usable knowledge.
Composite databases offer a holistic view with their integrated approach, allowing for complex queries and analyses drawn from multiple data types and sources.
Recognizing how these elements interplay and contribute to the broader bioinformatics landscape is vital to harnessing the full potential of these resources, driving innovation, and paving the way for discoveries in the field.
References
- Goudey B, Geard N, Verspoor K, Zobel J. Propagation, detection and correction of errors using the sequence database network. Brief Bioinform. 2022 Nov 19;23(6):bbac416. doi: 10.1093/bib/bbac416. PMID: 36266246; PMCID: PMC9677457.
- Bilofsky HS, Burks C. The GenBank genetic sequence data bank. Nucleic Acids Res. 1988 Mar 11;16(5):1861-3. doi: 10.1093/nar/16.5.1861. PMID: 3353225; PMCID: PMC338181.
- The UniProt Consortium. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017 Jan 4;45(D1):D158-D169. doi: 10.1093/nar/gkw1099. Epub 2016 Nov 29. Erratum in: Nucleic Acids Res. 2018 Mar 16;46(5):2699. PMID: 27899622; PMCID: PMC5210571.
- Gligorijević V, Pržulj N. Methods for biological data integration: perspectives and challenges. J R Soc Interface. 2015 Nov 6;12(112):20150571. doi: 10.1098/rsif.2015.0571. PMID: 26490630; PMCID: PMC4685837.
- Zarayeneh N, Ko E, Oh JH, Suh S, Liu C, Gao J, Kim D, Kang M. Integration of multi-omics data for integrative gene regulatory network inference. Int J Data Min Bioinform. 2017;18(3):223-239. doi: 10.1504/IJDMB.2017.10008266. Epub 2017 Oct 3. PMID: 29354189; PMCID: PMC5771269.
- Lee B, Zhang S, Poleksic A, Xie L. Heterogeneous Multi-Layered Network Model for Omics Data Integration and Analysis. Front Genet. 2020 Jan 28;10:1381. doi: 10.3389/fgene.2019.01381. PMID: 32063919; PMCID: PMC6997577.
- Collins FS, Fink L. The Human Genome Project. Alcohol Health Res World. 1995;19(3):190-195. PMID: 31798046; PMCID: PMC6875757.
- Joosten RP, te Beek TA, Krieger E, Hekkelman ML, Hooft RW, Schneider R, Sander C, Vriend G. A series of PDB related databases for everyday needs. Nucleic Acids Res. 2011 Jan;39(Database issue):D411-9. doi: 10.1093/nar/gkq1105. Epub 2010 Nov 11. PMID: 21071423; PMCID: PMC3013697.
- Ara T, Kodama Y, Tokimatsu T, Fukuda A, Kosuge T, Mashima J, Tanizawa Y, Tanjo T, Ogasawara O, Fujisawa T, Nakamura Y, Arita M. DDBJ update in 2023: the MetaboBank for metabolomics data and associated metadata. Nucleic Acids Res. 2023 Nov 16:gkad1046. doi: 10.1093/nar/gkad1046. Epub ahead of print. PMID: 37971299.
- Yuan D, Ahamed A, Burgin J, Cummins C, Devraj R, Gueye K, Gupta D, Gupta V, Haseeb M, Ihsan M, Ivanov E, Jayathilaka S, Kadhirvelu VB, Kumar M, Lathi A, Leinonen R, McKinnon J, Meszaros L, O'Cathail C, Ouma D, Paupério J, Pesant S, Rahman N, Rinck G, Selvakumar S, Suman S, Sunthornyotin Y, Ventouratou M, Vijayaraja S, Waheed Z, Woollard P, Zyoud A, Burdett T, Cochrane G. The European Nucleotide Archive in 2023. Nucleic Acids Res. 2023 Nov 13:gkad1067. doi: 10.1093/nar/gkad1067. Epub ahead of print. PMID: 37956313.
- Raney BJ, Barber GP, Benet-Pagès A, Casper J, Clawson H, Cline MS, Diekhans M, Fischer C, Navarro Gonzalez J, Hickey G, Hinrichs AS, Kuhn RM, Lee BT, Lee CM, Le Mercier P, Miga KH, Nassar LR, Nejad P, Paten B, Perez G, Schmelter D, Speir ML, Wick BD, Zweig AS, Haussler D, Kent WJ, Haeussler M. The UCSC Genome Browser database: 2024 update. Nucleic Acids Res. 2023 Nov 11:gkad987. doi: 10.1093/nar/gkad987. Epub ahead of print. PMID: 37953330.
- Redaschi, N., & UniProt Consortium. (2009). UniProt in RDF: Tackling Data Integration and Distributed Annotation with the Semantic Web. Nature Precedings. https://doi.org/10.1038/npre.2009.3193.1
- Burns, G., Krallinger, M., Cohen, K. et al. (2009). Biocuration Workflow Catalogue. Nat Prec. https://doi.org/10.1038/npre.2009.3250.1
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003 Jul 1;31(13):3784-8. doi: 10.1093/nar/gkg563. PMID: 12824418; PMCID: PMC168970.
- Andrew Yates, Wasiu Akanni, M. Ridwan Amode, Daniel Barrell, Konstantinos Billis, Denise Carvalho-Silva, Carla Cummins, Peter Clapham, Stephen Fitzgerald, Laurent Gil, Carlos García Girón, Leo Gordon, Thibaut Hourlier, Sarah E. Hunt, Sophie H. Janacek, Nathan Johnson, Thomas Juettemann, Stephen Keenan, Ilias Lavidas, Fergal J. Martin, Thomas Maurel, William McLaren, Daniel N. Murphy, Rishi Nag, Michael Nuhn, Anne Parker, Mateus Patricio, Miguel Pignatelli, Matthew Rahtz, Harpreet Singh Riat, Daniel Sheppard, Kieron Taylor, Anja Thormann, Alessandro Vullo, Steven P. Wilder, Amonida Zadissa, Ewan Birney, Jennifer Harrow, Matthieu Muffato, Emily Perry, Magali Ruffier, Giulietta Spudich, Stephen J. Trevanion, Fiona Cunningham, Bronwen L. Aken, Daniel R. Zerbino, Paul Flicek, Ensembl 2016, Nucleic Acids Research, Volume 44, Issue D1, 4 January 2016, Pages D710-D716, https://doi.org/10.1093/nar/gkv1157
- InterPro in 2022 Typhaine Paysan-Lafosse, Matthias Blum, Sara Chuguransky, Tiago Grego, Beatriz Lázaro Pinto, Gustavo A Salazar, Maxwell L Bileschi, Peer Bork, Alan Bridge, Lucy Colwell, Julian Gough, Daniel H Haft, Ivica Letunić, Aron Marchler-Bauer, Huaiyu Mi, Darren A Natale, Christine A Orengo, Arun P Pandurangan, Catherine Rivoire, Christian J A Sigrist, Ian Sillitoe, Narmada Thanki, Paul D Thomas, Silvio C E Tosatto, Cathy H Wu, Alex Bateman, Nucleic Acids Research (2022), gkac993, PMID: 36350672
- Deutsch EW, Orchard S, Binz PA, Bittremieux W, Eisenacher M, Hermjakob H, Kawano S, Lam H, Mayer G, Menschaert G, Perez-Riverol Y, Salek RM, Tabb DL, Tenzer S, Vizcaíno JA, Walzer M, Jones AR. Proteomics Standards Initiative: Fifteen Years of Progress and Future Work. J Proteome Res. 2017 Dec 1;16(12):4288-4298. doi: 10.1021/acs.jproteome.7b00370. Epub 2017 Sep 15. PMID: 28849660; PMCID: PMC5715286.
- Karsch-Mizrachi I, Takagi T, Cochrane G; International Nucleotide Sequence Database Collaboration. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2018 Jan 4;46(D1):D48-D51. doi: 10.1093/nar/gkx1097. PMID: 29190397; PMCID: PMC5753279.
