Protein Sequence Databases
UniProt
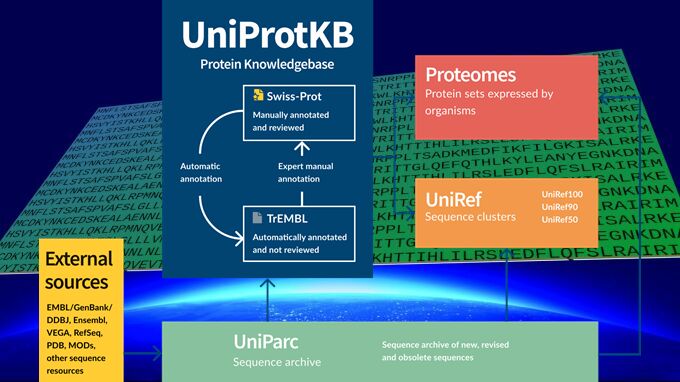
Image credit: EMBL-EBI Training (modified bakground) (CC BY 4.0)
Learning objectives:
- Have Basic understanding of the UniProt.
Overview
UniProt is the primary hub for comprehensive protein information, encompassing extensive data, and is freely accessible. Each database housed within UniProt has a distinct purpose, providing varied viewpoints on protein sequences and functions. The interconnected nature of these databases enables effortless exploration of protein data, facilitating seamless navigation across various levels of organization and annotation.
UniProt, a collaborative effort led by the UniProt Consortium involving the European Bioinformatics Institute (EMBL-EBI), the SIB Swiss Institute of Bioinformatics, and the Protein Information Resource (PIR), serves as a valuable resource for the scientific community. It offers a comprehensive repository of freely accessible protein sequence and functional information.
In response to the escalating number of fully sequenced genomes, researchers are actively working to understand the proteins encoded by these genomes, impacting fields such as biology, medicine, and biotechnology. UniProt plays a pivotal role by providing an up-to-date and thorough collection of protein data. This resource streamlines scientific discovery, saving researchers time and effort in data collection and organization.
UniProt's practical applications include retrieving information about specific proteins, comparing protein sequences, and mapping identifiers between external databases and UniProtKB. With almost 100 contributors engaged in database curation, software development, and user support tasks, UniProt is a collaborative hub facilitating scientific endeavors.
UniProtKB (UniProt Knowledgebase)
The UniProt Knowledgebase (UniProtKB) is a resource for obtaining functional protein details. Within UniProtKB, a specialized subset of entries forms the Proteomes dataset. This dataset focuses explicitly on collecting proteins predicted to be expressed by organisms with fully sequenced genomes, allowing researchers to delve into the proteomes of various organisms, providing valuable insights into their protein-level function and organization.
UniProtKB's primary sources of protein sequences include:
- Translations of coding sequences (CDS) from DDBJ/ENA/GenBank databases.
- Sequences directly derived from protein structures deposited in the Protein Data Bank (PDB).
- Sequences contributed by Ensembl and RefSeq databases.
- Sequences obtained from direct submissions to UniProtKB or extracted from the scientific literature.
However, not all protein sequences are incorporated into UniProtKB. The following types are excluded but are archived in UniParc (Box 1):
- Small fragments (less than eight amino acids in length).
- Synthetically generated sequences.
- Most non-germline immunoglobulins and T-cell receptors.
- Most sequences originate from patent applications.
- Pseudogenes.
- Sequences from redundant proteomes. See "Reducing proteome redundancy." (This link takes you to UniProt.)
As the UniProt Consortium's primary focus, the UniProt Knowledgebase is a meticulously curated protein database featuring two distinct sections: UniProtKB/Swiss-Prot and UniProtKB/TrEMBL.
Box 1. UniProt Archive (UniParc)
UniParc, short for UniProt Archive, is a comprehensive and non-redundant database of protein sequences. It complements the UniProt Knowledgebase (UniProtKB).
What it is:
- Massive: It contains the most publicly available protein sequences worldwide, including those not found in UniProtKB.
- Non-redundant All identical sequences are merged into one record, regardless of their source or species, eliminating repetition.
- Archive: It includes sequences even if they're no longer in their original databases.
- Cross-referenced: Each record links to the source databases for further information.
Think of it as:
- A vast reservoir of protein sequences: While UniProtKB is like a carefully curated library, UniParc is like a treasure trove containing everything, even less-studied or uncharacterized sequences.
- A tool for comprehensive sequence searches: By searching UniParc, you're searching all its source databases simultaneously, increasing your chances of finding relevant sequences.
- A resource for historical data: You can access even outdated sequences, which are valuable for studies tracking protein evolution or changes over time. Sequence version numbers are incremented every time the underlying sequence changes.
What it doesn't have:
- Detailed annotations: Unlike UniProtKB, UniParc only stores the protein sequence, and annotation and functional information can be accessed through the linked source databases.
- Strict quality control: While most sequences are from reliable sources, some might be less rigorously annotated or validated.
UniProtKB/Swiss-Prot: The Gold Standard of Protein Databases
UniProtKB/Swiss-Prot is the crown jewel of the UniProt Knowledgebase, a comprehensive resource for protein sequence and functional information. It's widely considered the gold standard in protein databases, offering researchers a wealth of curated, high-quality data on hundreds of thousands of proteins.
What makes UniProtKB/Swiss-Prot special?
- Expertly curated: Unlike other databases that rely on automated annotation, trained biologists meticulously curate Swiss-Prot entries, ensuring the accuracy and reliability of the information, making it ideal for critical research studies.
- Richly annotated: Each Swiss-Prot entry is a wealth of knowledge, packed with details like:
- Protein sequence: An amino acid sequence of a protein.
- Function: A detailed description of the protein's role in the cell or organism.
- Domains and motifs: Identify functional and structural units within the protein sequence.
- Post-translational modifications: How the protein is modified after being synthesized, affecting its function.
- References: UniProtKB/Swiss-Prot has links to the scientific literature for further exploration.
- Non-redundant: Swiss-Prot removes redundancy, meaning you won't find the same protein sequence listed multiple times for different reasons, making navigating and finding the information you need easier.
- Cross-linked: Swiss-Prot entries link to numerous external databases, providing easy access to additional resources like protein structures, interaction networks, and gene expression data. See an example Swiss-Prot record of: Histone H2B.10 on Swiss-Prot.
As you can see, the entry is packed with detailed information, including sequence, function, domains, post-translational modifications, and references.
Why use UniProtKB/Swiss-Prot?
Researchers use Swiss-Prot for a variety of purposes, including:
- Identifying and characterizing proteins: Finding information about specific proteins, such as their function, structure, and interactions with other molecules.
- Comparative analysis: Comparing proteins from different species to understand their evolution and functional differences.
- Drug discovery: Identifying potential drug targets by studying the function and structure of proteins in disease processes.
- Developing new tools and technologies: Using protein information to design new diagnostic tests, biomaterials, and therapeutic agents.
UniProtKB/Swiss-Prot is a precious resource for individuals involved in protein-related work. It provides a vast amount of precise, trustworthy, and well-structured information and is widely recognized as the benchmark for protein databases.
UniProtKB/TrEMBL: A Treasure Trove of Protein Sequences
UniProtKB/TrEMBL, a section of the UniProt Knowledgebase, is a vast and valuable resource for protein sequence data. Unlike its manually curated counterpart, UniProtKB/Swiss-Prot, TrEMBL focuses on unreviewed, computationally analyzed protein sequences, making it a powerhouse for exploring the ever-expanding universe of protein information, with over 251 million entries as of December 2023.
Key characteristics of UniProtKB/TrEMBL
UniProtKB/TrEMBL has several key characteristics that make it a valuable resource for protein research. Firstly, it offers a comprehensive collection of protein sequences from various organisms, including bacteria, archaea, eukaryotes, and viruses. This diversity allows researchers to explore proteins across various biological domains conveniently.
Additionally, UniProtKB/TrEMBL enhances each entry with computational annotation, incorporating information from automated analysis tools. These annotations include predictions related to protein function, structure, post-translational modifications, and other crucial features. Moreover, the database is regularly updated with new sequences and improved annotations, ensuring users can access the most up-to-date protein knowledge.
Lastly, UniProtKB/TrEMBL promotes open access, providing free data for download and exploration, benefiting researchers worldwide.
A breakdown of the information found in a typical TrEMBL entry
- Accession number: A unique identifier for a protein sequence.
- Protein name and description:
- Organism: The source organism of the protein sequence.
- Sequence features: Details about the amino acid composition, motifs, and domains present in the sequence.
- Function: Predicted or experimentally determined functions of the protein.
- Gene ontology (GO) terms: Standardized annotations describing the protein's molecular function, biological process, and cellular location.
- Cross-references: Links to other protein databases and resources for further investigation.
- Literature references: Citations to relevant scientific publications for deeper understanding.
TrEMBL's value as a research tool
TrEMBL is a valuable research tool that you can use for several purposes. Firstly, it can help in generating hypotheses by exploring proteins that are not well-characterized and identifying potential research targets.
Secondly, you can use it for comparative analysis by comparing protein sequences across different species to understand evolutionary relationships and functional conservation.
Thirdly, TrEMBL's rich annotations can be used for data mining to help discover patterns and trends in protein function and evolution.
Finally, TrEMBL can also be used to develop computational tools by training and testing algorithms for protein sequence analysis and prediction.
Despite its wealth of information, it's important to remember that TrEMBL entries are unreviewed. The computational annotations may not always be accurate, and experimental validation is recommended for critical research applications.
UniProt Reference Cluster (UniRef)
A Beacon of Order in the Protein Sequence Landscape
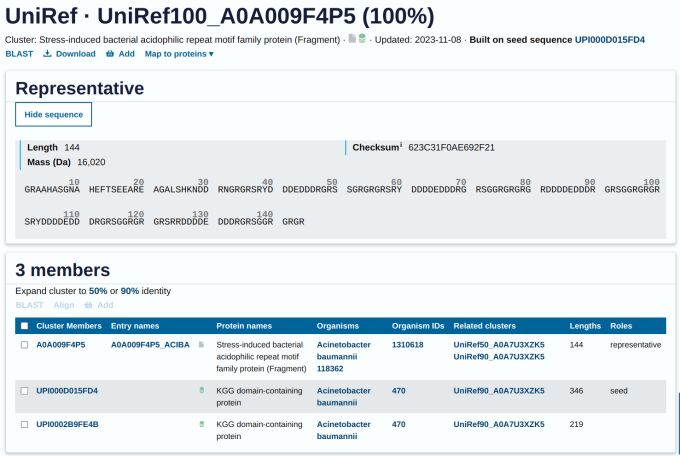
UniRef is a reliable and well-organized resource for protein sequence data. It is curated by the UniProt Consortium, which clusters protein sequences based on similarity, making it easier for researchers to analyze and explore protein data. This comprehensive resource offers a streamlined protein analysis and exploration approach, saving researchers time and effort.
UniRef's strength lies in its ability to eliminate redundancy within protein databases. UniRef clusters are generated through hierarchical clustering by grouping highly similar sequences into distinct clusters. By doing so, UniRef significantly reduces the search space, enabling researchers to navigate the vast protein landscape with remarkable speed and precision. There are three tiers of clusters, each serving a specific level of sequence identity.
- UniRef100: This category contains identical sequences and fragments with at least 11 amino acid residues, demonstrating the remarkable precision of nature's protein-folding mechanism.
- UniRef90: Stepping into this domain, we encounter sequences sharing a 90% identity threshold and an 80% overlap, their melodies echoing with the whispers of close evolutionary kinship.
- UniRef50: Descending further, we find distant relatives, bound by a minimum 50% identity and 80% overlap, their shared motifs hinting at a common ancestral origin, albeit obscured by the tides of evolutionary divergence.
UniRef's significance goes beyond mere data compaction, as its clustered architecture provides researchers with a powerful toolset. One essential capability is the unraveling of protein families. By identifying clusters containing related sequences, UniRef sheds light on the intricate tapestry of protein evolution, unveiling functional relationships and ancestral connections.
Another valuable function is the ability to pinpoint conserved domains. These ubiquitous modules, responsible for vital protein functions, often extend beyond individual sequence variations. UniRef facilitates the identification of these conserved domains, contributing to a deeper understanding of protein structure and function across diverse species.
UniRef also expedites homology searches, transforming the typically laborious task of finding proteins similar to a query sequence into a swift and efficient process. Researchers can achieve remarkable speed and accuracy by focusing their search within relevant clusters.
Moreover, UniRef is a valuable tool for constructing phylogenetic trees. It clusters sequences together, which helps researchers reconstruct the evolutionary history of protein families. By analyzing the distribution of sequences within and across clusters, you can map out the branching patterns of evolution, revealing how proteins are related to each other and their ancestral origins.
File Formats
Swiss-Prot and TrEMBL records use a flatfile format that is structured and easy to read. Computer applications can quickly parse this format. The flatfile format is a tabular text format that organizes information about proteins in a clear and structured way, making it both readable for humans and easily processed by computers. Each entry in the flatfile format contains specific information sections such as protein names, functions, references, and other relevant details—this format often exports and exchanges protein data between different bioinformatics tools and databases. See a sample record in the Appendix
The flatfile records have distinct features that make them easy to navigate and retrieve specific information. They are organized based on keywords, with each line starting with a two-letter keyword followed by relevant data. This structure allows users to find the information they need quickly.
Common keywords and their contents
- ID: Unique identifier for the protein (e.g., P01308, a web view for Insulin. See a flatfile example in the Appendix.)
- AC: Accession number, a stable identifier that doesn't change over time
- DT: Date of creation or last modification
- DE: Description of the protein's function
- GN: Gene name(s) associated with the protein
- OS: Organism(s) in which the protein is found
- OG: Organelle(s) in which the protein is located
- SQ: Protein sequence itself, in single-letter amino acid code
- FT: Feature table listing various protein features (e.g., domains, sites, modifications)
- DR: Database cross-references, linking to related information in other databases
- RN: References to scientific publications
- CC: Comment lines that provide extra information or explanations
- * : Star lines highlight important information or warnings
Other File Formats
Swiss-Prot provides two formats for distributing records: Plain Text (.txt) and (.dat) Format - Compressed Format Optimized for Computer Processing.
Humans can easily read the plain text format intentionally created for viewing and editing protein-related information. The format provides structured details such as names, functions, annotations, and references, making it a popular choice for manual curation and extracting specific information for analysis.
The Swiss-Prot/TrEMBL format (.dat) streamlines computer processing and storage, enhancing efficiency. It simplifies parsing and data extraction for computational tools and programs and optimizes the format for computational analysis. Therefore, it is an excellent option for bioinformatics, database queries, and large-scale data processing applications.
Flatfile format differences in the significant biological sequence repositories
Swiss-Prot/TrEMBL and GenBank/DDBJ are two important repositories for biological sequence data with differing data formats. Manual curators annotate the Swiss-Prot database, and automatic generators annotate the TrEMBL database using a flat-file format focusing on protein sequences and functional information. The format includes two-letter keywords: ID, AC, DT, DE, GN, OS, OG, SQ, FT, DR, RN, CC, and *.
The GenBank/DDBJ uses the ASN.1 format to store all biological sequences, such as DNA, RNA, and proteins, and the level of detail in the annotation of the sequences may vary based on the submission. The format includes sections such as LOCUS, DEFINITION, ACCESSION, FEATURES, and ORIGIN. For more information on the data format in nucleotide sequence databases, please refer to The Data Format In Nucleotide tutorial.
There are notable differences between the two formats. The Swiss-Prot/TrEMBL's flat-file format is simple, easy to read, and human-readable. In contrast, GenBank/DDBJ's ASN.1 format is flexible and suitable for complex data structures. In terms of annotation, Swiss-Prot/TrEMBL emphasizes manual curation and comprehensive annotation. On the other hand, GenBank/DDBJ encompasses both curated and minimally annotated entries. Swiss-Prot/TrEMBL primarily concentrates on protein sequences, while GenBank/DDBJ includes a broader spectrum of sequence types.
Swiss-Prot, TrEMBL, and GenBank/DDBJ have independently evolved, resulting in a variance in data formats. This variance is due to optimizing formats to suit specific data types and research questions within each database, which their historical origins have shaped.
References
-
Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006 Jan 1;34(Database issue):D187-91. doi: 10.1093/nar/gkj161. PMID: 16381842; PMCID: PMC1347523.
-
Lussi YC, Magrane M, Martin MJ, Orchard S; UniProt Consortium. Searching and Navigating UniProt Databases. Curr Protoc. 2023 Mar;3(3):e700. doi: 10.1002/cpz1.700. PMID: 36912607.
-
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, Poux S, Bougueleret L, Xenarios I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol Biol. 2016;1374:23-54. doi: 10.1007/978-1-4939-3167-5_2. PMID: 26519399.
-
Pundir S, Magrane M, Martin MJ, O'Donovan C; UniProt Consortium. Searching and Navigating UniProt Databases. Curr Protoc Bioinformatics. 2015 Jun 19;50:1.27.1-1.27.10. doi: 10.1002/0471250953.bi0127s50. PMID: 26088053; PMCID: PMC4522465.
-
Alpi E, Griss J, da Silva AW, Bely B, Antunes R, Zellner H, Ríos D, O'Donovan C, Vizcaíno JA, Martin MJ. Analysis of the tryptic search space in UniProt databases. Proteomics. 2015 Jan;15(1):48-57. doi: 10.1002/pmic.201400227. PMID: 25307260; PMCID: PMC4298651.
-
Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89-112. doi: 10.1007/978-1-59745-535-0_4. PMID: 18287689.
-
Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007 May 15;23(10):1282-8. doi: 10.1093/bioinformatics/btm098. PMID: 17379688.
- Redaschi, N., Consortium, U. UniProt in RDF: Tackling Data Integration and Distributed Annotation with the Semantic Web. Nat Prec (2009). 10.1038/npre.2009.3193.1.
Appendix
A truncated Swiss-Prot flatfile
ID INS_HUMAN Reviewed; 110 AA.
AC P01308; Q5EEX2;
DT 21-JUL-1986, integrated into UniProtKB/Swiss-Prot.
DT 21-JUL-1986, sequence version 1.
DT 08-NOV-2023, entry version 272.
DE RecName: Full=Insulin;
DE Contains:
DE RecName: Full=Insulin B chain;
DE Contains:
DE RecName: Full=Insulin A chain;
DE Flags: Precursor;
GN Name=INS;
OS Homo sapiens (Human).
OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia;
OC Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae;
OC Homo.
OX NCBI_TaxID=9606;
RN [1]
RP NUCLEOTIDE SEQUENCE [GENOMIC DNA].
RX PubMed=6243748; DOI=10.1038/284026a0;
RA Bell G.I., Pictet R.L., Rutter W.J., Cordell B., Tischer E., Goodman H.M.;
RT "Sequence of the human insulin gene.";
RL Nature 284:26-32(1980).
...
CC -!- FUNCTION: Insulin decreases blood glucose concentration. It increases
CC cell permeability to monosaccharides, amino acids and fatty acids. It
CC accelerates glycolysis, the pentose phosphate cycle, and glycogen
CC synthesis in liver.
CC -!- SUBUNIT: Heterodimer of a B chain and an A chain linked by two
CC disulfide bonds (PubMed:25423173). {ECO:0000269|PubMed:25423173}.
...
DR EMBL; V00565; CAA23828.1; -; Genomic_DNA.
DR EMBL; M10039; AAA59173.1; -; Genomic_DNA.
DR EMBL; J00265; AAA59172.1; -; Genomic_DNA.
DR EMBL; X70508; CAA49913.1; -; mRNA.
DR EMBL; L15440; AAA59179.1; ALT_SEQ; Genomic_DNA.
DR EMBL; AY899304; AAW83741.1; -; mRNA.
DR EMBL; AY138590; AAN39451.1; -; Genomic_DNA.
DR EMBL; BT006808; AAP35454.1; -; mRNA.
DR EMBL; CH471158; EAX02488.1; -; Genomic_DNA.
DR EMBL; BC005255; AAH05255.1; -; mRNA.
DR EMBL; AJ009655; CAA08766.1; -; Genomic_DNA.
DR CCDS; CCDS7729.1; -. [P01308-1]
DR PIR; A93222; IPHU.
...
KW 3D-structure; Alternative splicing; Carbohydrate metabolism;
KW Cleavage on pair of basic residues; Diabetes mellitus;
KW Direct protein sequencing; Disease variant; Disulfide bond;
KW Glucose metabolism; Hormone; Pharmaceutical; Reference proteome; Secreted;
KW Signal.
FT SIGNAL 1..24
FT /evidence="ECO:0000269|PubMed:14426955"
FT PEPTIDE 25..54
FT /note="Insulin B chain"
FT /id="PRO_0000015819"
FT PROPEP 57..87
FT /note="C peptide"
FT /id="PRO_0000015820"
...
SQ SEQUENCE 110 AA; 11981 MW; C2C3B23B85E520E5 CRC64;
MALWMRLLPL LALLALWGPD PAAAFVNQHL CGSHLVEALY LVCGERGFFY TPKTRREAED
LQVGQVELGG GPGAGSLQPL ALEGSLQKRG IVEQCCTSIC SLYQLENYCN
//
