Construction of substitution matrices
part II
PAM matrices
The approach behind the construction of PAM substitution matrices differs from the construction of BLOSUM matrices, although the goal is the same, which is the building of a scoring system to identify homologous sequences, yielding a specific score for each type of match and substitution, based on empirical observations of substitutions occurred in known sequences. The best way to accomplish this is to study identified homologous sequences, but the catch is that we only can study present-day sequences and consequently we cannot directly observe the history of how sequences evolved.
To infer a history of substitutions, we must compare similar sequences between different species, but yet another obstacle is that since we cannot see back in history, how do we know that substitution of, e.g., Tyrosine is direct to Tryptophan? There could have been several substitutions earlier in the history. The following could have happened: The first Tyrosine substitution was to Phenylalanine after that back to Tyrosine then to Histidine and finally to Tryptophan again. Consequently, when inferring substitution probabilities, we must be confident that several substitutions did not occur in between the one we observe; otherwise our probabilities are incorrect.
Figure 1 illustrates an imaginary evolutionary tree, where an early ancestor's gene duplication gives rise to genes (1) and (2) that also went through duplication and in turn evolved to two pairs (3-4) and (5-6). A comparison of these pairs gives the following three substitutions: H-K, Q-K, T-S, and T-Y. If we were to compare all sequences (3 to 6) together, we would get two additional substitutions, namely H-Q and S-Y.
Importantly note that no substitutions happened between any of the genes from 3 to 6. The actual substitutions are K to H between (1) and (3), T to S between (1) and (4), G to Q between (2) and (5), and L to Y between (2) and (6). We can see this in our imaginary tree, but in reality, we cannot observe ancestral gene sequences - only present-day sequences.
Consequently, in an attempt to overcome this obstacle, and construct the BLOSUM matrices, the approach of Henikof and Henikof took was division the families of clusters into sub-clusters by their percentage of similarity to reduce multiple contributions to amino acid pair frequencies, whereas Dayhoff and colleagues used phylogenetic trees to guide the construction of PAM matrices.
Dayhoff and colleagues reasoned that two separate processes result in an observable mutation; first, a mutation in a template sequence, in our tree genes (1) and (2), and second, to be accepted by natural selection, i.e., mutations that were not deleterious thus surviving to the present day and called them accepted point mutations, APMs. However, they later changed the name to PAM, perhaps because it was easier to pronounce. They used data from closely related sequences from 34 families and grouped them into 71 evolutionary trees and observed in total 1,572 changes.
So why would this approach work? We just stated that it is possible only to observe present-day sequences. So, how can we construct real evolutionary trees in the first place? Real evolutionary trees depict a relationship between species, in this case between genes, and can give an evolutionary distance between genes that we can observe today, arranged hierarchically. This hierarchical arrangement yields information about which present-day genes have a common ancestor and an evolutionary distance to it. For example sequence (1) is the common ancestor to the genes (3) and (4) in Figure 1. However, in general, we have no information about what happened in the paths from a common ancestor evolving to the present-day species (Figure 1, genes 3 and 4). Nevertheless, by minimizing the distance to a common ancestor, we at the same time minimize the number of unknowns. In other words, if the distance between, e.g., (1) and (4) was close to zero, we could directly observe the substitution T to S. Note that in this case gene (1) would be a present-day gene that we can observe and although an ancestral gene it is still alive. Despite this, we can never be sure that the observed genes are direct ancestors.
Several different ways to measure an evolutionary distance between sequences exist, but they all necessarily relate to a degree of similarity alternatively a difference between genes. Consider an extreme of two genes that are identical then the evolutionary distance is zero.
For this reason, Dayhoff and colleagues only constructed trees consisting of sequences that were at least 85% identical and the ancestral sequences represented by trees even more similar. To illustrate the approach, see Figure 2 where we tabulated all accepted point mutations from our imaginary tree in Figure 1. The assumption is that the likelihood of substitution XY is the same as YX; thus, the method does not allow measurements over evolutionary distances, but the distances are in PAMs. More about this a bit later. Note that Dayhoff also included the differences between genes (1) and (2) as accepted point mutations.
Dayhoff's, and colleagues' original count multiplied by 10 is in Figure 3. Note that they never observed 35 of all the possible substitutions, because of the low degree of divergence between the observed sequences, at most 15% and perhaps some of the substitutions are lethal and did not survive to be observed. Other substitution counts vary from three up to 831. This uneven variability is due to similarities and differences between amino acid chemistries (See amino acid groups). For example, (E-D) Glutamic acid and Aspartic acid both are negatively charged, (S-A) Serine and Alanine both are tiny, whereas Phenylalanine (F) is hydrophobic, Glutamine (Q) and Glutamic acid (E) are polar and charged, i.e., hydrophilic and consequently the table doesn't contain any recorded substitutions for these amino acids.
Another explanation is that the genetic code is redundant, meaning that DNA can code for the same amino acid with a varied number of different codons. For example, six different codons code for Serine and consequently a mutation from TCT to TCC in the DNA sequence does not alter the coded amino acid, since both codons code for Serine. Tryptophan and Methionine are on the other hand result of a single codon. We can see that Serine is substituted relatively often to the contrary of Methionine which is also a result of a single codon. However, Methionine is also the start codon marking the beginning of a gene sequence in genomes; therefore, a mutation in this codon is more likely to be deleterious than a mutation in Serine codon.
Dayhoff and colleagues also calculated the relative mutability of amino acids, which is a count of the number of times each amino acid is substituted divided by the number of times it occurs in an observed interval. For example, if we have the aligned sequences AGLL and AGAV the relative mutability calculation is as follows:
A G L L
A G A V
Amino acids: A G L V
Changes: 1 0 2 1
Frequency of occurrence: 3 2 2 1
Relative mutability: 0.33 0 1 1
These frequencies need to be normalized and are in the table of normalized frequencies of the amino acids that underlie the computation of the accepted point mutation data in Figure 3.
Making of the mutation probability matrix for the evolutionary distance of one PAM (PAM1)
Recall that to calculate a probability of observed substitutions \(P_{ab}\); we need to know the frequency of each type of substitutions \( fs_{a} \) and \( fs_{b} \), so that \( P_{ab} = fs_{a} fs_{b} \). Moreover, to calculate the probability of seeing a particular substitution or score \( S_{a,b} \), we need the frequencies of occurrences \(f\) of each amino acid and calculate the probability of observing them together by chance, which is \( f_{a}f_{b} \), so that the probability to observe a nonrandom substitution is \( S_{a,b} = \frac{ P_{ab} }{ f_{a}f_{b} } \) (eq. 1).
Dayhoff and colleagues used the accepted point mutation matrix (Figure 3) and the relative mutability data to get the values corresponding to \( P_{ab} \) and \( f_{a}f_{b} \) in equation 1.
The way to calculate mutation probabilities for PAM matrices is slightly different from the BLOSUM approach for the reason that it is possible to extrapolate them to a desired evolutionary distances measured in PAMs, starting from the PAM1 matrix (figure 4). It consists of sequences that on average are 1% different, that is about one mutation per 100 amino acids which is a distance of one PAM. The sum of each column is one, meaning that each amino acids substitution probabilities with another amino acid within a column sum to one as it should be that all the probabilities within a sample space must sum to one. For example, observing a mutation in a site containing Alanine is one minus the probability of not observing a mutation (1 - 0.9867 = 0.013 = 1.33%) or the other way round, the sum of all the substitution probabilities in the Alanine column, except Alanine-Alanine ( \(P_{R}+P_{N}+P_{D}+ ...+P_{V} = 1.33% \) ). However, the sum of rows is not necessarily equal to one.
First, we calculate the nondiagonal values \( M_{ij} \) as follows:
\[ M_{ij} = \frac{ m_{j} A_{ij} }{ \sum_{i} A_{ij} } \: \: (2) \]where \( A_{i} \), is a value of the accepted point mutation matrix (Figure 3), and \( m_{i} \) is the mutability of the \(j\)th aminoacid in Table Relative Mutabilities of the Amino Acids. Don't give in quite yet; we explain the equation in detail below.
To complete the PAM1 matrix, we still need the diagonal values and calculate them as follows:
\[ M_{ij} = 1 - m_{i} \: \: (3)\]Specific calculations
Along the way, we construct two different types of PAM matrices first a mutation probability matrix that gives the probability of an amino acid (j) be replaced by an amino acid (i) after some specific evolutionary time. The second matrix is a relational log-odds matrix based on the mutation probability matrix.
The point accepted mutations table (Figure 3) is the starting point, and we note that only the left half has data in it. However, since we assume that the mutations are directionally identical, for example, mutation of A to R is the same as the mutation from R to A; we add the right half by copying the left to the right side to make the matrix dimensionally square and set the diagonal values to zeros (red color) for the time being. Then we calculate the sums of columns and rows, although we only need the column sums, having both is a way to check we copied correctly. The sums are the number of times A, R, N, ..., V mutated, for example, in column one A mutated to some other amino acid 3,644 times and in the second column R mutated to some other 1,112 times. We don't yet know how many times they were not mutated, but we find out shortly. The accepted point mutation matrix thus becomes this:
Table 1.
A R N D C Q E G H I L K M F P S T W Y V SUM
A 0 30 109 154 33 93 266 579 21 66 95 57 29 20 345 772 590 0 20 365 3644
R 30 0 17 0 10 120 0 10 103 30 17 477 17 7 67 137 20 27 3 20 1112
N 109 17 0 532 0 50 94 156 226 36 37 322 0 7 27 432 169 3 36 13 2266
D 154 0 532 0 0 76 831 162 43 13 0 85 0 0 10 98 57 0 0 17 2078
C 33 10 0 0 0 0 0 10 10 17 0 0 0 0 10 117 10 0 30 33 280
Q 93 120 50 76 0 0 422 30 243 8 75 147 20 0 93 47 37 0 0 27 1488
E 266 0 94 831 0 422 0 112 23 35 15 104 7 0 40 86 31 0 10 37 2113
G 579 10 156 162 10 30 112 0 10 0 17 60 7 17 49 450 50 0 0 97 1816
H 21 103 226 43 10 243 23 10 0 3 40 23 0 20 50 26 14 3 40 30 928
I 66 30 36 13 17 8 35 0 3 0 253 43 57 90 7 20 129 0 13 661 1481
L 95 17 37 0 0 75 15 17 40 253 0 39 207 167 43 32 52 13 23 303 1428
K 57 477 322 85 0 147 104 60 23 43 39 0 90 0 43 168 200 0 10 17 1885
M 29 17 0 0 0 20 7 7 0 57 207 90 0 17 4 20 28 0 0 77 580
F 20 7 7 0 0 0 0 17 20 90 167 0 17 0 7 40 10 10 260 10 682
P 345 67 27 10 10 93 40 49 50 7 43 43 4 7 0 269 73 0 0 50 1187
S 772 137 432 98 117 47 86 450 26 20 32 168 20 40 269 0 696 17 22 43 3492
T 590 20 169 57 10 37 31 50 14 129 52 200 28 10 73 696 0 0 23 186 2375
W 0 27 3 0 0 0 0 0 3 0 13 0 0 10 0 17 0 0 6 0 79
Y 20 3 36 0 30 0 10 0 40 13 23 10 0 260 0 22 23 6 0 17 513
V 365 20 13 17 33 27 37 97 30 661 303 17 77 10 50 43 186 0 17 0 2003
SUM 3644 1112 2266 2078 280 1488 2113 1816 928 1481 1428 1885 580 682 1187 3492 2375 79 513 2003
To turn the accepted mutation counts into a mutation probability matrix, we need help from the amino acid mutability table and use it to compute the background probabilities for each column corresponding to the \( \sum_{i} A_{ij} \) in Equation 2. We start by diving each mutation count sum by the corresponding mutability of amino acid as follows:

The results above are the respective background frequencies for each amino acid, i.e., the probabilities of observing them mutated by chance, and next, and they still need to be normalized though. We do this by first computing the sum of the background frequencies (= ~418.24) and then divide each value by the sum, resulting in the following normalized vector of frequencies (Table 2):
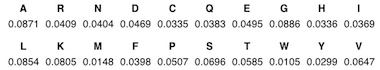
After that, we need to compute the total probability space given both the amino acid mutabilities and background probabilities by taking a dot product of the vectors. A dot product is multiplying each entry in the first vector by a corresponding entry in the second vector and summing the products. In this case, 100 x 0.871 + 65 x 0.0409 + 134 x 0.0404, ..., 74 x 0.0647 = ~75.15.
To get the final total scaled mutation probability space, we divide each entry in the mutability vector by 7,515 as follows:
A: 100 / 7,515 = 0.0133
R: 65 / 7,515 = 0.0086
N: 134 / 7,515 = 0.0178
...
...
...
V: 74/7,515 = 0.00985.
Now we can compute the diagonal probabilities as in Equation 3:
A: 1 - 0.0133= 0.9867
R: 1 - 0.0086 = 0.9914
N: 1 - 0.0178 = 0.9822
...
...
...
V: 1 - 0.00985 = 0.9902
To reiterate, the sum of probabilities in each column is one; thus, one minus the total mutation probability space equals the probability that A, R, N,..., V respectively do not mutate.
To complete the mutation probability matrix PAM 1, we only need to fill in the diagonal values. Of course, first we need to scale them also by dividing the final mutation probability space by the sums of each column to give us the scale factors for each column as follows:
A: 0.0133 / 3,644 = ~ 0.00000365R: 0.0086 / 1,112 = ~0.00000778
N: 0.0178 / 2,266 = ~0.00000787
...
...
...
V: 0.00985 / 2,003 = ~0.00000492
Finally, we use these scale factors to compute the final probabilities corresponding to \( m_{j}A_{ij} \) in Equation 2. We multiply the scale factors by each corresponding mutation count row-wise in the accepted square point mutation matrix in Table 1 above. Also, we multiply by 10,000 to get the original look as follows:
First column:A: 0.00000365 x 30 x 10,000 = 1
R: 0.00000365 x 109 x 10,000 = 4
N: 0.00000365 x 154 x 10,000 = 6
...
...
...
V: 0.00000365 x 365 x 10,000 = 13
For the column two, we use the scale factor 0.00000778 and for the third column 0.00000787 and so on until the matrix is complete. Below, is the complete probability matrix PAM 1 (Figure 5).
Importantly note that this mutation probability matrix is not symmetric. However, we explore next how to create any PAM matrix by extrapolating from PAM1. In contrary to the mutation matrix, these matrices are symmetric.
Extrapolation of PAM1 mutation matrix
PAM1 matrix based on 99% identical sequences represent the evolutionary distance where one percent of amino acids have changed and thus are optimal to compare 99% identical sequences. However, most of the time it is more interesting to compare sequences that are less identical.
It is possible to use a scaling factor to scale the substitution frequencies to the desired evolutionary distance, just by multiplying with a value of lambda ( \( \lambda \) ), which we can add to Equation 2: \[ M_{ij} = \frac{ \lambda m_{j} A_{ij} }{ \sum_{i} A_{ij} } \: \: (4) \]
However, this approach does not take into account multiple substitutions that can occur. Therefore, a better approach is to use matrix multiplication. In other words, multiply PAM1 by itself many times to arrive at some particular evolutionary distance measure in PAMs. The accuracy of the multiplication results depends on the accuracy of PAM1.
How do we multiply matrices? The primary method to multiply matrices is best to show by an example:
| A | B | C | J | K | L | |||
| D | E | F | X | M | N | O | = | |
| G | H | I | P | Q | R |
| [AJ + BM + CP] [AK + BN + CQ] [AL + BO + CR] |
| [DJ + EM + FP] [DK + EN + FQ] [DL + EO + FR] |
| [GJ + HM + IP] [GK + HN + IQ] [GL + HO + IR] |
[Click on the image to toggle zoom ◱ ]
Note though that the rule of matrix multiplication makes the order of multiplication to matter! We multiply matrices from left to right, although when multiplying PAM1 with PAM1 the order does not make any difference since they are both the same. In contrary, PAM2 x PAM5, for example, is not the same as PAM5 x PAM2. So, in which order should we multiply PAM1 by itself to get, e.g., PAM 3? The answer is, we always keep PAM1 on the left side, thus, e.g., PAM1 x PAM249 = PAM250.
Look at it like this: The cell A-A in the PAM2 matrix is the probability that A does not mutate or the probability we observe A-A. Therefore, to get the value for A-A after PAM1 x PAM1 or one PAM1 period after PAM1, we need first to calculate the following:
- A. The probability that A stays A, which is (A-A) x (A-A).
- B. Calculate the probability that any of the other amino acids mutate to A, which is the sum of all probabilities on the first row (R-A, N-A, D-A,..., V-A), except the first, A-A.
- C. Calculate the probability that A mutates to any of the other amino acids, which are all the probabilities on the first column (A-R, A-N, A-D,..., A-V), except A-A.
So the value of A-A in the PAM2 matrix is then the sum of the probabilities A+B+C. Precisely according to the matrix multiplication rule. PAM1 x PAM1 = PAM2 is shown in the Figure 6.
[Click on the image to toggle zoom ◱ ]
This way Dayhoff and colleagues modeled protein evolution and it is called the Markov chain model, where the current state only depends on the previous state. In this case, the previous probability that an amino acid mutates to some other amino acid, but we need to take into account the whole probability space and thus apply the matrix multiplication rule. Introduction to Markov chain: simplified, is an excellent simplified explanation of the Markov chain.
How does this model behave when we keep on multiplying the matrices? We see that with every evolutionary distance step the amino acid frequencies from being one in PAM0 slowly converge to that of their background frequencies with increasing distances. If this were true in real life, all the species would be dead by now, since all the gene sequences would be random and thus unlikely to have any function. It is a little bit like leaving a hot cup of coffee on a table, and after a while, the coffee becomes the same temperature as the room. That is the second law of thermodynamics in action.
[Click on the image to toggle zoom ◱ ]
Figure 7 shows the effect of the amino acid frequencies converging to background frequencies with increasing distance. However, if we don't go to extremes, this bias is not extraordinarily substantial, and PAM matrices up to PAM250 are still in use even though it represents an evolutionary distance where amino acids have mutated 2.5 times, i.e., 250% change. Despite this, we can observe that 20% are identical. See table 3 for identity percentages for other distances.
Next, we explore how to construct log-odds matrices based on PAM mutation matrix. Continue to the next page.