Protein Sequence Databases
NCBI Protein Sequence Database

Learning objectives:
- Have Basic understanding of the NCBI Protein Sequence Database.
Introduction
The NCBI Protein database is a comprehensive repository of protein sequences from diverse origins. These records adhere to the same format as GenBank Nucleotide records (see The Data Format In Nucleotide Sequence Databases), and a substantial portion originates from annotated coding regions (CDS) found in GenBank, RefSeq (see the tutorial on RefSeq), and Third-Party Annotation (TPA) sequences.
In addition to these primary sources, the Protein database incorporates data from various other protein databases, notably SwissProt, the Protein Information Resource (PIR), the Protein Research Foundation (PRF), and the Protein Data Bank (PDB). Below are some insights into the contributing resources that enrich the NCBI Protein database.
GenBank Nucleotide Translation
The GenPept database, housing protein sequences translated from GenBank's CDS, plays a pivotal role. While not an official NCBI release, GenPept is meticulously maintained and synchronized with each new GenBank release, providing valuable protein sequence information.
RefSeq Translation
RefSeq transcripts contribute protein records, known as RefSeq proteins (reference proteins). Distinguished by accession numbers prefixed with 'NP' for experimentally generated transcripts and 'XP' for model transcripts, these RefSeq proteins are non-redundant and enjoy robust support from the NCBI (see our tutorial on RefSeq).
UniProtKB/Swiss-Prot
A substantial contributor, UniProtKB/SwissProt, stands out as a curated protein sequence database (See tutorial on UniProt ). Its well-annotated, less redundant sequences are seamlessly integrated with other databases. In 2002, the European Bioinformatics Institute (EBI), the Swiss Institute of Bioinformatics (SIB), and the Protein Information Resource (PIR) united to form the UniProt consortium.
Protein Information Resource ( PIR)
PIR is a significant contributor, offering an integrated public resource of protein sequences and supporting information for genomic and proteomic research. Its Protein Sequence Database ( PIR-PSD) contains annotated sequences spanning the entire taxonomic range.
Protein Research Foundation (PRF)
PRF contributes substantially to the NCBI Protein database, particularly as a source for predicted protein sequences reported in scientific literature.
Protein Data Bank (PDB)
The PDB is the most extensive database for protein sequences and their three-dimensional structures. Biologists and biochemists worldwide submit data obtained through X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy, making it a cornerstone in protein research.
Keyword Searches Using The Web Interface - A Quick Look
Let's do an example keyword search. Our "Searching and Querying Sequence Databases" tutorial contains more advanced methods. You can access the protein database at the URL www.ncbi.nlm.nih.gov/protein/. For advanced queries, refer to our tutorials on "Searching Databases." includes some indexed fields of the Protein database.
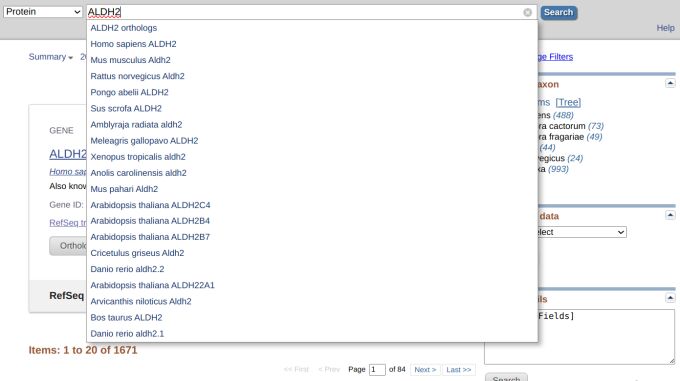
First, we enter a simple query, a gene name, ALDH2, and nothing else. We see a drop-down menu where you can choose a species from the drop-down menu list (Figure 1).
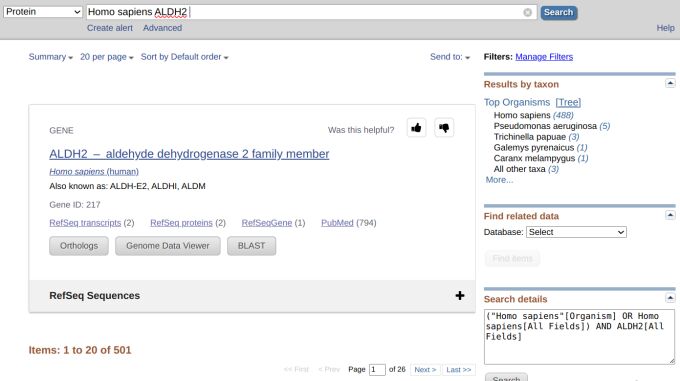
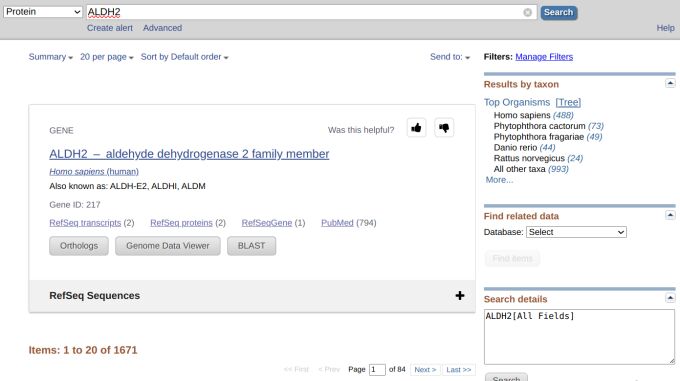
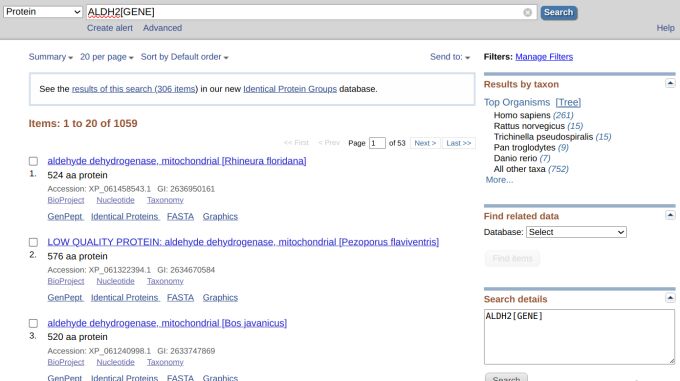
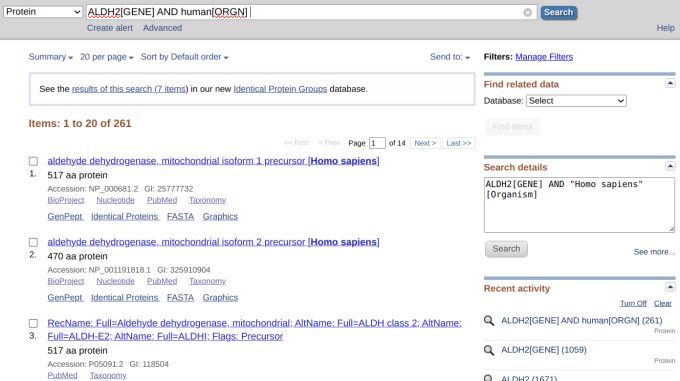
However, we got 501 results, which we did not want (Figure 2). But still, there were much fewer results if we didn't select from the drop-down menu and queried using the gene name (Figure 3). Let's refine our search by inputting "ALDH2[GENE]" since GENE is an indexed field in the protein database. This time, however, we got 1,059 results (Figure 4).
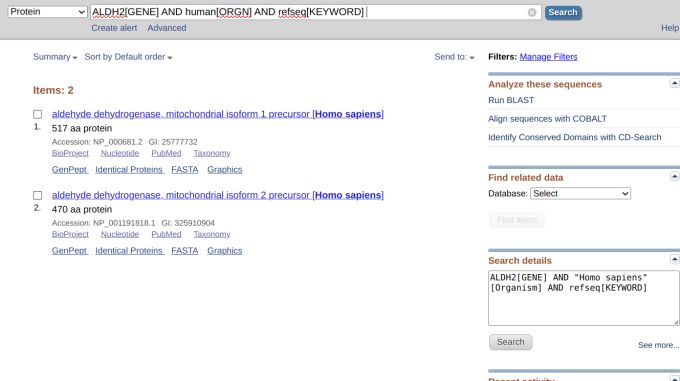
We will not give up, though, and add "AND human[ORGN]" so that the search becomes "ALDH2[GENE] AND human[ORGN]." Now, we see that the number of results has shrunk to 261. We can still refine the search even more by adding "AND refseq[KEYWORD" so that the query becomes "ALDH2[GENE] AND human[ORGN] AND refseq[KEYWORD]." This time, we got only two results, which was what we might have expected because RefSeq is a non-redundant database (Figure 6).
It is also good to know that for other types of searches, we might want to exclude something and can use the "NOT' operator, for example, insulin "NOT receptor[KEYWORD]." Also, we can use a wild card "*" for partial matches, such as "cyclin*." If you have the accession number of the protein, you can directly search using it, for example, NP_000178 (for insulin receptor).
Index Fields In NCBI Protein Database
| Description | Short Field | Description | Short Field |
|---|---|---|---|
| Accession | ACCN | Publication date | PDAT |
| All fields | ALL | Primary organism | PORG |
| Assembly | ASSM | Properties | PROP |
| Author | AUTH | Protein name | PROT |
| Division | DIV | Sequence length | SLEN |
| Filter | FILT | SeqID string | SQID |
| Gene name | GENE | Strain | STRN |
| BioProject | GPRJ | Title | TITL |
| Keyword | KYWD | UID | UID |
| Organism | ORGN | Text word | WORD |
References
-
NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018 Jan 4;46(D1):D8-D13. doi: 10.1093/nar/gkx1095. PMID: 29140470; PMCID: PMC5753372.
