129 Free Genome-wide Association Study (GWAS) Tools - Software and Resources
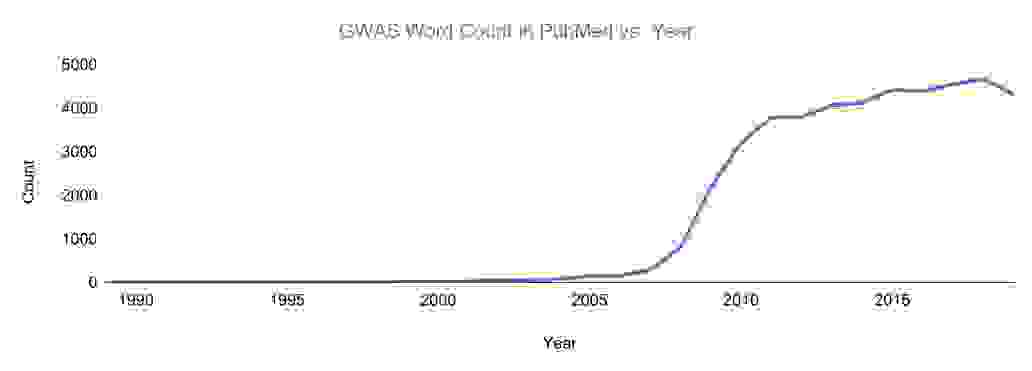
Selected published GWAS review papers are at the bottom of this page.
The advanced search function is under maintenance and coming up shortly. We apologize the inconvenience.
-
AlphaDrop_beta
- Description : AlphaDrop beta is a tool to simulate genomic selection and genome-wide association study (GWAS) data. It can simulate sequence data, SNP data, pedigrees, QTL effects, and breeding values. The AlphaDrop algorithm drops simulated haplotypes through a pedigree using MaCS. Available by request from John Hickey.
-
Altools
- Description : Uses the BWA/SAMtools/VarScan pipeline to call SNPs and indels, and the dnaCopy algorithm for genome segmentation to identify copy number variations. Uses insert size information to detect large deletions.
-
aSPU
- Description : aSPU is a tool to test association of gene- and pathway-trait in genome-wide association study (GWAS) summary data.
-
Bayenv
- Description : Software implementing a Bayesian method that uses a set of markers to estimate a pattern of covariance in allele frequencies between different populations. The estimation is used as a null model for testing individual SNPs.
-
BiForce Toolbox
- Description : BiForce Toolbox is a web-based tool for the analysis of pair-wise epistasis of quantitative and disease traits in genome-wide association study (GWAS) data.
-
BigTop
- Description : BigTop is a tool to visualize Manhattan plots in 3D using a virtual reality framework.
-
BioBin
- Description : BioBin is a tool to investigate the rare variant burden in genetic trait studies and the natural distribution of rare variants in ancestral populations aimed for analyzes and hypothesis testing. The BioBin algorithm generates bins based on various features, such as regulatory and evolutionary conserved regions, genes, and pathways. BioBin uses information stored in the LOKI database, the Library of Knowledge Integration.
-
BLINK
- Description : BLINK (Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway) is a tool for genome-wide association studies (GWAS) to identify genes controlling human diseases and agricultural traits. The algorithm uses Bayes and linkage disequilibrium information.
-
BWMR
- Description : BWMR is a tool to infer causal effects on phenotypes on outcome in genome-wide association study (GWAS) data. The BWMR algorithm uses a Bayesian weighted Mendelian randomization (BWMR) and a variational expectation-maximization (VEM) method.
-
CERENKOV
- Description : CERENKOV is a tool for the identification of functional noncoding single nucleotide polymorphism (SNP) in loci identified by genome-wide association studies (GWAS). The CERENKOV algorithm uses data-space geometric features and the xgboost classifier.
-
cit
- Description : cit is a tool to test hypotheses mediation analysis. It uses the causal inference test (CIT) based on hypothesis testing.
-
CPBayes
- Description : CPBayes is a tool for the meta-analysis of cross-phenotype genetic associations. The CPBayes algorithm uses summary data from multiple phenotypes to assess the evidence of the aggregate-level pleiotropic association and for the estimation of additional trait associations at the risk locus.
-
EMMAX
- Description : EMMAX (Efficient Mixed-Model Association eXpedited) is a tool for testing association mapping considering the sample structure in genome-wide association studies (GWAS). The EMMAX algorithm uses a variance component approach that can analyze GWAS datasets within hours.
-
EPIQ
- Description : EPIQ is a tool to detect epistasis in quantitative GWAS. The EPIQ algorithm uses metric embedding and random projections to eliminate the need to exhaustively test all SNP pairs.
-
Evoker
- Description : A tool to visualize genotype cluster plots and provides a storage.
-
famFLM
- Description : famFLM is a tool for the region-based association analysis using functional linear models. The famFLM algorithm also has a function for the association test between quantitative traits and multiple regional genetic variants. The famFLM tool is part of the FREGAT package.
-
FarmCPU
- Description : FarmCPU (Fixed and random model Circulating Probability Unification) is a tool for genome-wide association analyses (GWAS). The algorithm uses the general linear model framework. Following single-marker regression, FarmCPU assigns the data into bins and classifies a set of optimal markers as covariates in the subsequent iteration.
-
FaST-LMM
- Description : FaST-LMM (Factored Spectrally Transformed Linear Mixed Models) is a tool for large genome-wide association studies (GWAS).
-
FORGE
- Description : FORGE is a tool for the discovery of cell-specific enrichments in genome-wide association study (GWAS) associated single nucleotide polymorphisms (SNPs). FORGE also visualizes enrichment summaries.
-
FunciSNP
- Description : A tool to integrate functional non-coding data sets with genetic association studies for identification of regulatory SNPs.
-
G2P
- Description : G2P (A Genome-Wide-Association-Study Simulation Tool for Genotype Simulation, Phenotype Simulation, and Power Evaluation) is a tool to simulate genotypes for the genome-wide association studies (GWAS). The G2P can simulate genotype data, phenotype data and evaluate the statistical power.
-
GAPIT
- Description : GAPIT (Genome Association and Prediction Integrated Tool) is a tool for genome-wide association study (GWAS) and for genome prediction or selection. The GAPIT algorithm uses The Mixed Linear Model (MLM).
-
garfield
- Description : garfield is an R tool that uses genome-wide association study (GWAS) data together with annotations to find appropriate phenotypes. The garfield algorithm uses greedy pruning of GWAS data and assigns annotations based on overlaps of functional information. It also computes Fold Enrichment and uses permutations testing to evaluate them and computes major allele frequency (MAF), LD proxies, and distances to the nearest TSS.
-
GBOOST
- Description : GBOOST is a tool to detect gene-gene interactions in genome-wide case-control studies (GWAS). The GBOOST tool runs in GPU (CUDA/Nvidia) and does Boolean based screening and testing. CUDA Driver : 2.3.
-
gboosting
- Description : gboosting is a tool to boost gradients and analysis of the false discovery rate in survival analyses for genome-wide association studies (GWAS).
-
gcatest
- Description : gcatest is a tool to test genome-wide associations (GWAS) with arbitrarily complex population structures. The gcatest algorithm uses a genotype-conditional association test (GCAT), a new method.
-
GCORE-sib
- Description : GCORE-sib is a tool for the analysis of genome-wide gene interaction. The GCORE-sib algorithm can test interactions of a large number of single-nucleotide polymorphism (SNP) in genome-wide association studies (GWAS).
-
GDT
- Description : GDT (generalized disequilibrium test) is a tool for genome-wide association studies (GWAS). The GDT algorithm has functions for generalized family-based association tests for dichotomous traits using the genotype discrepancies of all discordant corresponding pairs in evaluating association within a family.
-
GEMMA
- Description : GEMMA (genome-wide efficient mixed-model association) is a tool for testing association in genome-wide association study (GWAS) data. The GEMMA algorithm computes exact Wald statistics and p-values.
-
GenoWAP
- Description : GenoWAP is a tool to prioritize signals, integrate functionals annotation, and GWAS test statistics in genome-wide association study (GWAS) results.
-
GIGSEA
- Description : GIGSEA (Genotype Imputed Gene Set Enrichment Analysis) is a tool to analyze imputed genotypes. The GIGSEA algorithm uses a combination of genome-wide association study (GWAS) summary statistics and eQTL to deduce differential gene expression and to examine enrichment for gene sets.
-
GISPA
- Description : GISPA (Gene Integrated Set Profile Analysis) is a tool to determine specified gene sets with a similar molecular profile.
-
GLOGS
- Description : GLOGS (Genome-wide LOGistic mixed model/Score test) is a tool for the improvement of power in genome-wide association studies (GWAS) in related individuals.
-
GMDR
- Description : GMDR is a tool to identify gene-gene and gene-environment Interactions for complex traits. The GMDR tool can use continuous, count, dichotomous, polytomous nominal, ordinal, survival and multivariate, and unrelated case-control, family-based and pooled unrelated and family samples, and can adjust covariates.
-
GPA
- Description : GPA (Genetic analysis incorporating Pleiotropy and Annotation) is a tool for the prioritization of genome-wide association studies (GWAS) results using pleiotropy information and annotation data. The GPA algorithm has functions for fitting models and hypothesis testing the associated SNPs.
-
gPLINK
- Description : gPLINK is a GUI tool for the PLINK command-line tool (see links) to perform several main operations. It also makes it possible for the computation itself to be located on another server.
-
graph-GPA
- Description : graph-GPA is a tool to prioritize the genome-wide association study (GWAS) results and to examine multiple phenotypes. The graph-GPA algorithm uses a hidden Markov random field method.
-
GraphAT
- Description : GraphAT is a tool to test associations between multiple sources. The algorithm uses graph theory and makes edge permutation and node label permutation tests.
-
GW-SEM
- Description : Genome-Wide Structural Equation Modeling. A method to test the association of a SNP with multiple phenotypes or a latent construct on a genome-wide basis using a diagonally weighted least squares (DWLS) estimator for four common SEMs; a one-factor model, a one-factor residuals model, a two-factor model, and a latent growth model.
-
GWAPP
- Description : GWAPP is a tool for genome-wide association studies (GWAS) in Arabidopsis thaliana.
-
GWAS catalog
- Description : GWAS catalog is a catalog of publicly available, manually curated, and published genome-wide association study (GWAS) data, containing over 100k single-nucleotide polymorphisms (SNPs) and trait associations.
-
GWAS Pipeline
- Description : GWAS Pipeline is a pipeline tool for genome-wide association analysis (GWAS). The GWAS pipeline can filter, create a kinship matrix, covariate files, run EMMAX, computes Manhattan and QQ plots. The GWAS has functions for computing a summary of the most significant SNPs with calculated allele effects. Requirements: Python v2.6-v2.7, NumPy, Plink v1.9, and R.
-
GWAS3D
- Description : GWAS3D is a web-based tool for the computation of the probability of genetic variants that associate with regulatory pathways and respective diseases or traits. The GWAS3D algorithm integrates chromatin state, functional genomics, sequence motif, and conservation information with GWAS data or a variant list. See also GWAS4D server (links).
-
GWAS4D
- Description : GWAS4D is a web-based tool for the analysis of genome-wide association study (GWAS) data. GWAS4D is an updated version GWAS3D and has algorithms for variant prioritization, incorporation of tissue-specific epigenetic data, transcriptional regulatory motifs, processing of Hi-C data, non-coding variant functional annotations, interactive visualization of SNP target interaction.
-
gwascat
- Description : gwascat is a tool to represent and model EMBL-EBI GWAS catalog.
-
GWASdb
- Description : GWASdb is a database of genome variants that are identified using genome-wide association studies (GWAS). The database is manually curated and includes annotations for genetic variants.
-
GWASpi
- Description : GWASpi (genome-wide association studies pipeline) is a tool to manage and analyze single-nucleotide polymorphism (SNPs) in genome-wide association studies (GWAS).
-
GWASTools
- Description : GWASTools is an R tool for quality control, analysis, and annotation of genome-wide association study (GWAS) data. The package stores data in NetCDF format to allow data sets that exceed the R memory limits.
-
GWAtoolbox
- Description : GWAtoolbox is a tool for quality control and handling of data files originating from genome-wide association studies (GWAS).
-
GWIZ-Rscript
- Description : GWIZ-Rscript is a tool to assess the performance of genome-wide association studies (GWAS) for the prediction of disease risk. The GWIZ-Rscript algorithm uses ROC curves and calculates the AUROC using GWAS summary-level data.
-
HaploView
- Description : HaploView is a tool to analyze and visualize LD haplotype maps. The HaploView includes functions for LD and haplotype block analysis, population frequency estimation, single SNP and haplotype association tests, permutation testing, Paul de Bakker's Tagger tag SNP selection algorithm, download of phased genotype data from HapMap, visualization, and plotting.
-
HiView
- Description : HiView is a tool to interactively browse Hi-C results for genome-wide association study GWAS variants.
-
i-GSEA4GWAS
- Description : i-GSEA4GWAS is a web-based tool for the identification of pathways and genes that are associated with specific traits in genome-wide association studies (GWAS) results. It uses NP label permutation to correct gene variation to decrease biases.
-
IGES
- Description : IGES (Gentic Analysis integrating individual level data and summary statistics) is a tool for the identification of risk variants and risk prediction in genome-wide association studies (GWAS). The IGES algorithm integrates individual level genotypes and summary statistics to increase statistical power in the analyses.
-
ImpG-Summary
- Description : A tool for genotype imputation using. The ImpG-Summary algorithm uses Gaussian imputation with summary association statistics.
-
iPat
- Description : iPat (Intelligent Prediction and Association Tool) is a Java tool for genome-wide association studies (GWAS). The graphica user interface allows drag and click specification of input data files, choosing input parameters and selection of models. iPat includes GAPIT, PLINK, FarmCPU, BLINK, rrBLUP and BGLR models.
-
IPGWAS
- Description : IPGWAS is a pipeline tool that combines genome-wide association study (GWAS) and quality control. Apart from quality control, IPGWAS includes functions for Manhattan and quantile-quantile plots, and format conversion for genetic analyses, genotype phasing, and imputation.
-
JASS
- Description : JASS (Joint Analysis of Summary Statistics) is a tool for the joint analysis of genome-wide association study (GWAS) data. The JASS algorithm includes, for example, the omnibus approach and many weighted sums of -score tests.
-
KGG
- Description : KGG is a tool package for genome-wide association studies(GWAS). The KGG includes
-
LAMPLINK
- Description : Detects statistically significant SNP combinations from genome-wide case-control data.
-
LDSC
- Description : LDSC is a tool to estimate heritability and genetic correlations in genome-wide association study (GWAS) dataset summary statistics. The LDSC algorithm uses linkage disequilibrium (LD) score regression among test statistics and LD to discriminate inflation from true polygenic signals and biases.
-
lme4
- Description : lme4 is a tool for fitting linear and generalized linear mixed-effects models. The lme4 algorithm presents models and components with S4 classes.
-
LocusZoom
- Description : LocusZoom (LocusZoom.js) is a tool for visualization of genome-wide association study (GWAS) scan results. LocusZoom uses LD information from HapMap, 1000 Genomes, gene information from the UCSC browser.
-
lodGWAS
- Description : lodGWAS is a tool for the analysis of biomarkers in genome-wide association studies (GWAS). The lodGWAS algorithm uses a parametric survival analysis technique to specifically analyze biomarkers restricted by a Limit of Detection (LOD).
-
MACH2QTL
- Description : MACH2QTL is a tool for QTL analysis of imputed dosages or posterior probabilities.
-
MACLEAPS
- Description : MACLEAPS is a tool to predict disease risk in genome-wide association studies (GWAS). The MACLEAPS algorithm uses support vector machines.
-
Manhattan
- Description : Manhattan is a tool to annotate, plot, and display genome-wide association study (GWAS) data.
-
Manhattan-Harvester
- Description : Manhattan-Harvester is a tool to detect peaks genome-wide association study (GWAS) summary data. The Manhattan-Harvester also computes quality scores and various parameters to characterize individual peaks.
-
martini
- Description : martini is a tool for the analysis of low power in genome-wide association studies (GWAS). The martini algorithm incorporates prior knowledge as a network where SNPs are the vertices.
-
Matapax
- Description : Matapax is a pipeline tool for genome-wide association studies (GWAS). It displays the results, candidate markers both in tabular and genome browser formats.
-
Merlin
- Description : Merlin (multipoint engine for rapid likelihood inference) is a tool for pedigree analysis. The Merlin algorithm uses sparse trees to represent gene flow in pedigrees and the tool is useful for LD and association analyses, ibd, kinship, haplotyping, detection of errors, and simulations.
-
metaCCA
- Description : metaCCA is a tool for the meta-analysis of genome-wide association study (GWAS) data based on summary statistics and using canonical correlation analysis.
-
METAINTER
- Description : A tool to perform meta-analysis of summary statistics of related studies, such as single-marker association tests.
-
Metal
- Description : Metal is a tool for meta-analysis of genomewide association scans.
-
MetaSKAT
- Description : MetaSKAT (SNP-set Sequence Kernel Association Test) is an R tool that provides functions for Meta-analysis Burden tests using summary statistics.
-
miRNASNP2
- Description : miRNASNP2 is a database for the selection of single nucleotide polymorphism (SNP) by GWAS and miRNA expression data. The miRNASNP2 also includes tools for the analysis of the impact of new variants on miRNA:mRNA binding, multiple filters to prioritize functional SNP selection and to predict the consequence of miRNA related to a particular SNP.
-
MSS
- Description : MSS (maximal segmental score procedure) is a tool for the analysis of genome-wide association study (GWAS) data. The MSS algorithm uses region-specific empirical p-values for the identification of genomic segments and scores them using Fisher's p-value in combination with locus-specific significance levels.
-
MULTIPOW
- Description : MULTIPOW is a tool for the computation of statistical power for joint and replication-based analysis of general multi-stage genetic association studies.
-
mvBIMBAM
- Description : mvBIMBAM is a tool for multivariate association analysis. The mvBIMBAM algorithm uses Bayesian statistics and is an adaptation of the BIMBAM tool (see links).
-
NAM
- Description : NAM is a tool to study associations in multiple populations. The NAM algorithm treats makers as random effects and uses a sliding-window method to prevent fitting the same markers multiple times. It also accounts for prior population stratification.
-
OmicABEL
- Description : OmicABEL is a tool for genome-wide association analysis (GWAS). The OmicABEL has two separate algorithms for single and multiple traits and uses a mixed-model.
-
OrdinalGWAS
- Description : OrdinalGWAS is a tool for genome-wide association studies (GWAS) for ordered categorical phenotypes.
-
PAPA
- Description : PAPA is a tool for the analysis of pleiotropic pathways using genome-wide association study (GWAS) summaries.
-
PARIS
- Description : PARIS (Pathway Analysis by Randomization Incorporating Structure) is a tool for the analysis of pathways to aggregate signals in genome-wide association studies (GWAS) results. The PARIS algorithm clusters SNPs into LD, LE groups, creates groups according to pathways and tests the significance by permutation testing. The download requires registering.
-
Pascal
- Description : Pascal (Pathway scoring algorithm) is a tool to score and analyze pathways in genome-wide association study (GWAS) data. The algorithm uses enrichment strategies for scoring
-
PBAT
- Description : A tool for family-based association studies. It can accomodate missing parental genotypes, pedigrees with missing genotypes, and analysis of single nucleotide polymorphism (SNPs).
-
PC-select
- Description : PC-select is a tool for the computation of association statistics in genome-wide association studies (GWAS). The PC-select algorithm uses a data-adaptive GRM to improve power and avoids confounding from population stratification.
-
PEPIS
- Description : PEPIS (Pipeline for estimating EPIStatic genetic effects) is a web-based tool for the estimation of polygenic Epistatic QTL mapping for genome-wide association studies (GWAS). The PEPIS algorithm uses a new linear mixed model and includes kinship matrix calculation, polygenic component analyses, and genome scanning for main and epistatic effects.
-
PExFInS
- Description : PExFInS (Post-GWAS Explorer for Functional Indels and SNPs) is a tool to analyze functional insertions and deletions (indels), and single-nucleotide polymorphism (SNPs) in genome-wide association study (GWAS) data. The PExFInS algorithm includes functions for Linkage disequilibrium (LD) analysis with genotyping data of SNPs and indels from the 1000 Genomes Project, Comparison of LD pattern of query variants, eQTL analysis, annotation using ANNOVAR.
-
PheGWAS
- Description : PheGWAS is a tool for the visualization of multiple genotypes as 3D Manhattan plot landscapes.
-
PheWAS
- Description : PheWAS (Phenome-wide association studies) is a tool for genome-wide association studies (GWAS). The method has the potential to identify therapeutic and adverse drug outcomes in electronic health record data.
-
PLATO
- Description : PLATO (PLatform for the Analysis, Translation, and Organization of large-scale data) is a tool that includes functions for quality control, filtering, and multiple analytics methods for genome-wide association study (GWAS) data. Requires login.
-
PLINK
- Description : PLINK is a popular and well-documented tool for the association and population-based linkage analyses for genome-wide association studies (GWAS). The PLINK package contains functions for data management, summary statistics for quality control, population stratification detection, association analysis, identity-by-descent estimation, and more. See also gPLINK, a GUI tool written in Java for PLINK.
-
powerGWASinteraction
- Description : powerGWASinteraction is a tool to compute the statistical power of gene-gene and gene-environment interactions in genome-wide association studies (GWAS).
-
PSESM
- Description : A tool for detecting SNP main effects and SNP-SNP interactions.
-
QCGWAS
- Description : QCGWAS is a tool for quality control of genome-wide association study (GWAS) results.
-
QCTOOL
- Description : QCTOOL is a tool to administrate and quality control data from genome-wide associations studies (GWAS). The QCTOOL algorithm has functions for computing variant, sample QC metrics, for filtering, merging datasets, format conversion, annotation, LD between variants, genotype comparison, relatedness, principal components, genetic risk predictor scores, Hardy-Weinberg Equilibrium test, and multiple ways to manipulate the datasets.
-
qMSAT
- Description : qMSAT (quality-based Multivariate Score Association Test) is a tool for multivariate association testing of rare variants. The qMSAT algorithm weights the scores by sequencing quality values.
-
QTDT
- Description : QTDT is a tool for the analysis of linkage disequilibrium (LD) of Quantitative and Discrete Traits. The QTDT has algorithms for exact p-values by permutation although when testing multiple linked polymorphisms, tests described by Allison (TDTQ5, 1997), Rabinowitz (1997), Monks et al. (1998), Fulker et al. (1999), and association models: Abecasis (2000), the models of Allison (1997, TDTQ5), Rabinowitz (1997), Monks (1998) and Fulker (2000).
-
Rainbow (Janssen R and D)
- Description : Rainbow (Janssen R and D) is a wrapper for Crossbow pipeline tool (see links) whole-genome sequencing analysis. It can process single nucleotide polymorphism (SNP) data for genome-wide association studies (GWAS).
-
RAISS
- Description : RAISS is a tool for the imputation of single-nucleotide polymorphisms (SNPs) in genome-wide association study (GWAS) summary data. The RAISS is suitable for multi-trait analyses.
-
RaMWAS
- Description : RaMWAS is a tool to analyze methylome-wide associations. RaMWAS includes algorithms for mutual analysis of methylation and genotype data.
-
regioneR
- Description : regioneR is a tool for the association analysis of genomic regions. The regioneR algorithm uses permutation tests.
-
RegScan
- Description : RegScan is a tool for the association analysis of allele frequencies and continuous traits in genome-wide association studies (GWAS). The RegScan algorithm uses linear regression to predict marker effects on continuous traits.
-
rqt
- Description : rqt is a tool for the meta-analysis of genome-wide association study (GWAS) data.
-
Scoary
- Description : Scoary is a tool to score associations phenotypes to pan-genome components. The Scoary algorithm considers population stratification with the least possible assumptions of evolutionary processes and outputs a list of genes sorted by strength of trait association.
-
seq2pathway
- Description : seq2pathway is a tool for the functional gene-set analysis of genomic loci in RNA-seq data sets. The seq2pathway algorithm assigns genes to the pathways and computes gene-level pathway scores.
-
SEQPower
- Description : SEQPower is a tool for the analysis of statistical power and the estimation of sample sizes in association studies
-
SIMreg
- Description : SIMreg is a tool for maker-set association analysis at gene, pathway, and exon levels to evaluate the etiological effects of genes in genome-wide association study (GWAS) or sequence data.
-
SKAT
- Description : SKAT is a tool for the evaluation of the cumulative effect of sequence variants by association tests. The SKAT algorithm combines tests of SNP sets and computes p-values and power vs. sample size to assist in experiment design.
-
SNPEVG
- Description : SNPEVG is a tool to view single nucleotide polymorphism (SNP) effects for genome-wide association studies (GWAS).
-
snpGeneSets
- Description : snpGeneSets is a tool to annotate the genome-wide association study (GWAS) data. The snpGeneSets tool includes functions for genomic mapping annotation of SNPs, genes, the functional annotation for gene sets, the bidirectional mapping between SNPs and genes, and genes and gene sets, calculation of gene effects, gene set enrichment analyses for the identification of pathways.
-
SNPRelate
- Description : SNPRelate is an R tool for computing relatedness and also provides principal component (PCA) analysis. The SNPRelate algorithm produces a binary format for single-nucleotide polymorphism (SNP) data in genome-wide association study (GWAS) data.
-
SNPStats
- Description : A web-based tool containing classes of statistical methods, extending snpMatrix package.
-
SNPsyn
- Description : SNPsyn is a tool
-
SNPTEST
- Description : SNPTEST is a tool to analyze a single SNP association in genome-wide association studies (GWAS). The SNPTEST algorithm includes functions for case-control phenotypes, single and multiple quantitative phenotypes Bayesian, Frequentist tests, condition using an arbitrary set of covariates and/or SNPs, and several methods for imputed SNPs.
-
SOLAR-eclipse
- Description : SOLAR-eclipse is a tool to analyze genetic variance. The SOLAR-eclipse algorithm includes functions for SNP association, quantitative genetic, and linkage analyses.
-
SPOT
- Description : SPOT is an online tool to prioritize single-nucleotide polymorphism (SNP) after a genome-wide association study. The SPOT tool combines information from biological databases to allow biologically relevant SNPs to have an increased priority.
-
SSEA
- Description : SSEA (SNP Set Enrichment Analysis) is a tool to analyze pathway enrichment in genome-wide association studies (GWAS). The SSEA algorithm first identifies representative SNPs using adaptive truncated product statistics, ranks the selected SNPs, and tests their significance using a weighted Kolmogorov-Smirnov test.
-
STEGO
- Description : STEGO (Similarity Test for Estimating Genetic Outliers) is a tool for the identification of genetic outliers caused by sub-structure and cryptic relationships in high-throughput genetic association studies.
-
SurvivalGWAS_SV
- Description : SurvivalGWAS_SV is a tool for the analysis of imputed genotypes in genome-wide association studies (GWAS) with "time-to-event" outcomes. The SurvivalGWAS_SV algorithm uses Cox proportional hazards or Weibull regression models. It can also accommodate multiple covariates and include SNP-covariate interplay effects.
-
TASSEL
- Description : A tool to evaluate trait associations, linkage disequilibrium, and evolutionary patterns.
-
traseR
- Description : An R package to analyze trait associated single nucleotide polymorphism (SNP) enrichment. Visualization of results.
-
treeWAS
- Description : treeWAS is a tool for genome-wide association studies (GWAS) for microbes. The treeWAS algorithm uses a clonal population structure and homologous recombination to increase the precision of the computation.
-
UNCcombo
- Description : UNCcombo is a tool for Likelihood-based complex trait association testing.
-
Variant Ranker
- Description : Variant Ranker is a web-based tool to rank and annotate variants in genome-scale. Variant Ranker includes several modules: 1. Variant Ranker: Single sample VCF/List of Variants, 2. Filtering: Multi-sample VCF/Case-Control Filtering, 3. Filtering: Result Explorer, 4. Network Analyser, and 5. SNPtoGene.
-
VSEAMS
- Description : VSEAMS (variant set enrichment analysis using multivariate sampling) is a pipeline tool for the analysis of genes and genomic intervals enrichment in genome-wide association study (GWAS) data. The VSEAMS algorithm computes p-values for a trait using GWAS summary statistics.
-
wtest
- Description : wtest is a tool for association testing of principal effects, pairwise and high order interactions in genome-wide association study (GWAS) data. The wtest also has functions for genome-wide and epigenome-wide testing of the cis-regulation of SNP and CpG sites.
-
XGR
- Description : XGR is a tool to evaluate genome-wide association study (GWAS) summary data and also quantitative trait loci (eQTL) data. XGR utilizes prior biological knowledge and relationships.
If you find errors, please report here: comments and suggestions.
Scientific Reviews on GWAS To top
Visscher, Peter M et al. “10 Years of GWAS Discovery: Biology, Function, and Translation.” American journal of human genetics vol. 101,1 (2017): 5-22. doi:10.1016/j.ajhg.2017.06.005 PMCID: PMC5501872.
Cantor, Rita M et al. “Prioritizing GWAS results: A review of statistical methods and recommendations for their application.” American journal of human genetics vol. 86,1 (2010): 6-22. doi:10.1016/j.ajhg.2009.11.017. PMCID: PMC2801749.
Gallagher, Michael D, and Alice S Chen-Plotkin. “The Post-GWAS Era: From Association to Function.” American journal of human genetics vol. 102,5 (2018): 717-730. doi:10.1016/j.ajhg.2018.04.002. PMCID: PMC5986732
Power, Robert A et al. “Microbial genome-wide association studies: lessons from human GWAS.” Nature reviews. Genetics vol. 18,1 (2017): 41-50. doi:10.1038/nrg.2016.132
Mooney, Michael A et al. “Functional and genomic context in pathway analysis of GWAS data.” Trends in genetics : TIG vol. 30,9 (2014): 390-400. doi:10.1016/j.tig.2014.07.004. PMCID: PMC4266582.
Jia, Peilin, and Zhongming Zhao. “Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives.” Human genetics vol. 133,2 (2014): 125-38. doi:10.1007/s00439-013-1377-1. PMCID: PMC3943795.
Hayes, Ben. “Overview of Statistical Methods for Genome-Wide Association Studies (GWAS).” Methods in molecular biology (Clifton, N.J.) vol. 1019 (2013): 149-69. doi:10.1007/978-1-62703-447-0_6
Huang, Qingyang. “Genetic study of complex diseases in the post-GWAS era.” Journal of genetics and genomics = Yi chuan xue bao vol. 42,3 (2015): 87-98. doi:10.1016/j.jgg.2015.02.001
Marigorta, Urko M et al. “Replicability and Prediction: Lessons and Challenges from GWAS.” Trends in genetics : TIG vol. 34,7 (2018): 504-517. doi:10.1016/j.tig.2018.03.005. PMCID: PMC6003860.
De, Rishika et al. “Bioinformatics challenges in genome-wide association studies (GWAS).” Methods in molecular biology (Clifton, N.J.) vol. 1168 (2014): 63-81. doi:10.1007/978-1-4939-0847-9_5
Cannon, Maren E, and Karen L Mohlke. “Deciphering the Emerging Complexities of Molecular Mechanisms at GWAS Loci.” American journal of human genetics vol. 103,5 (2018): 637-653. doi:10.1016/j.ajhg.2018.10.001. PMCID: PMC6218604.
Duncan, Laramie E et al. “How genome-wide association studies (GWAS) made traditional candidate gene studies obsolete.” Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology vol. 44,9 (2019): 1518-1523. doi:10.1038/s41386-019-0389-5. PMCID: PMC6785091.
Giral, Hector et al. “Into the Wild: GWAS Exploration of Non-coding RNAs.” Frontiers in cardiovascular medicine vol. 5 181. 17 Dec. 2018, doi:10.3389/fcvm.2018.00181. PMCID: PMC6304420.
Dourlen, Pierre et al. “Using High-Throughput Animal or Cell-Based Models to Functionally Characterize GWAS Signals.” Current genetic medicine reports vol. 6,3 (2018): 107-115. doi:10.1007/s40142-018-0141-1. PMCID: PMC6096908.
Duo Jiang and Miaoyan Wang. "Recent developments in statistical methods for GWAS and high-throughput sequencing association studies of complex traits." Biostatistics & Epidemiology, (2018) 2:1, 132-159, doi:10.1080/24709360.2018.1529346.
Berlanga-Taylor, Antonio J. “From Identification to Function: Current Strategies to Prioritise and Follow-Up GWAS Results.” Methods in molecular biology (Clifton, N.J.) vol. 1793 (2018): 259-275. doi:10.1007/978-1-4939-7868-7_15.
SECTIONS
TutorialsIn-house software
Blog
News
Find Jobs
History
Definition of Bioinformatics
ENCYCLOPEDIA
COVID-19
⚬ Timeline of outbreak
⚬ Blog
⚬ Scientific Facts
⚬ Daily statistics
⚬ News
TOOLS
1. SNP Tools
2. GWAS Tools
3. Genotype Imputation Tools
4. MSA tools
5. CNV tools
6. WGA tools
7. RNA-seq tools
8. Proteomics Tools
9. Nucleic Acid Structure Analysis Tools
10. Nucleic Acid Sequence Analysis Tools
11. Sequence Logo Tools
12. In-house Software Tools
Find thousands of Bioinformatics and Life Science software tools and databases in the newly launched
Ads
Ads
Ads
