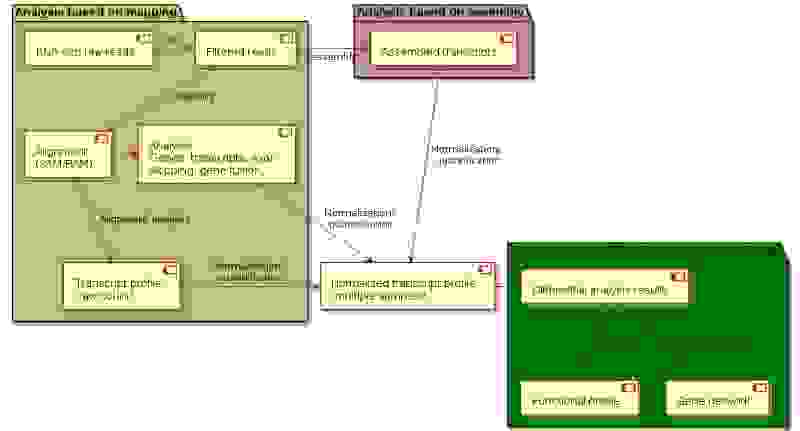
We have assigned each RNA-seq tool into categories by the computational task. The single-cell specific RNA-seq (scRNA-seq) tools are on the scRNA-seq collection page.
RNA-seq Pre-analysis Tools
Pre-analysis quality control of raw reads includes assessment of tolerable GC and k-mer contents, removal of sequence adaptors, PCR artifacts, and contaminations. The assessment of duplicates and sequencing errors. In addition, sequencing quality tends to decrease towards the 3' end of the reads; Thus, the reads must be trimmed to remove the low-quality ends
-
- Filtering (12 tools)
- Trimming (19 tools)
- Filtering and Trimming (13 tools)
- Reporting/Visualization (17 tools)
- Other Pre-analysis RNA-seq Tools (16 tools)
RNA-seq Core Analysis Tools
-
-
-
- Unstranded (4 tools)
- Stranded (11 tools)
- Quality Control (2 tools)
-
- Splice Aware (26 tools)
- Splice unaware (8 tools)
- Quality Control (15 tools)
-
-
- Union-exon Based (7 tools)
This method counts a read to belong to a gene if its alignment has adequate overlap with a 'union-exon.' A 'union-exon' consists of all overlapping exons of a gene. - Transcript Based (40 tools)
This method distributes reads between transcript isoforms of a gene. - Bacterial (4 tools)
In bacteria, genes do not contain introns; Thus, the genes do not have alternatively spliced transcript isoforms.
- Pre-processing DEA (4 tools)
To remove biases, normalization of data before DEA is a must to achieve reliable quantification of gene expression levels. For example, reads mapping on multiple locations can introduce bias. Many tools combine DEA analysis with a normalization step, but some tools solely perform normalization possibly combined diverse testing. - Parametric (42 tools)
Parametric tests assume certain defined population distributions from which the data originates. - Non-parametric (13 tools)
Non-parametric tests do not make any prior assumptions of population distributions. - Power analysis (11 tools)
We should do a power analysis to find out the number of samples required to achieve the desired level of obtaining statistical significance in the differential expression (DE) analyses. Ideally, we should do a power analysis prior to sample collection. Note, that, e.g., data normalization and multiple hypothesis testing affect the power analysis.
- Enrichment Analysis (GSEA), annotation, other (76 tools)
We perform gene set enrichment analysis to find out which genes are over-represented in an analysis set and thus potentially associated with a specific phenotype. - Comparison With Genome (2 tools)
RNA-seq Visualization and Other Analysis Tools
-
- Web-Based (17 tools)
- Stand-alone (22 tools)
-
- Gene fusion (8 tools)
- Pipelines (27 tools)
- Other (64 tools)
Scientific Review articles of RNA-seq
Geistlinger L, Csaba G, Santarelli M, Ramos M, Schiffer L, Turaga N, Law C, Davis S, Carey V, Morgan M, Zimmer R, Waldron L. "Toward a Gold Standard for Benchmarking Gene Set Enrichment Analysis." Brief Bioinform. 2020 Feb 6:bbz158. doi:10.1093/bib/bbz158
Wang, Liguo et al. “Measure transcript integrity using RNA-seq data.” BMC bioinformatics vol. 17 58. 3 Feb. 2016, doi:10.1186/s12859-016-0922-z
Ji, Fei, and Ruslan I Sadreyev. “RNA-seq: Basic Bioinformatics Analysis.” Current protocols in molecular biology vol. 124,1 (2018): e68. doi:10.1002/cpmb.68
Chatterjee, Aniruddha et al. “A Guide for Designing and Analyzing RNA-Seq Data.” Methods in molecular biology (Clifton, N.J.) vol. 1783 (2018): 35-80. doi:10.1007/978-1-4939-7834-2_3
Zhang, Hong et al. “Transcriptome Sequencing: RNA-Seq.” Methods in molecular biology (Clifton, N.J.) vol. 1754 (2018): 15-27. doi:10.1007/978-1-4939-7717-8_2
Li, Xing et al. “Quality control of RNA-seq experiments.” Methods in molecular biology (Clifton, N.J.) vol. 1269 (2015): 137-46. doi:10.1007/978-1-4939-2291-8_8
Rodríguez-García, Antonio et al. “RNA-Seq-Based Comparative Transcriptomics: RNA Preparation and Bioinformatics.” Methods in molecular biology (Clifton, N.J.) vol. 1645 (2017): 59-72. doi:10.1007/978-1-4939-7183-1_5
Love, Michael I et al. “RNA-Seq workflow: gene-level exploratory analysis and differential expression.” F1000Research vol. 4 1070. 14 Oct. 2015, doi:10.12688/f1000research.7035.1
Marco-Puche, Guillermo et al. “RNA-Seq Perspectives to Improve Clinical Diagnosis.” Frontiers in genetics vol. 10 1152. 12 Nov. 2019, doi:10.3389/fgene.2019.01152
Liang, Hanquan, and Erliang Zeng. “RNA-Seq Experiment and Data Analysis.” Methods in molecular biology (Clifton, N.J.) vol. 1366 (2016): 99-114. doi:10.1007/978-1-4939-3127-9_9
Baruzzo G, Hayer KE, Kim EJ, Di Camillo B, FitzGerald GA, Grant GR. Simulation-based comprehensive benchmarking of RNA-seq aligners. Nat Methods. 2017;14(2):135–139. doi:10.1038/nmeth.4106
Han H, Men K. How does normalization impact RNA-seq disease diagnosis?. J Biomed Inform. 2018;85:80–92. doi:10.1016/j.jbi.2018.07.016
Todd, Erica V et al. “The power and promise of RNA-seq in ecology and evolution.” Molecular ecology vol. 25,6 (2016): 1224-41. doi:10.1111/mec.13526
Chen, Geng et al. “Characterizing and annotating the genome using RNA-seq data.” Science China. Life sciences vol. 60,2 (2017): 116-125. doi:10.1007/s11427-015-0349-4
Tarazona, Sonia et al. “Differential expression in RNA-seq: a matter of depth.” Genome research vol. 21,12 (2011): 2213-23. doi:10.1101/gr.124321.111
Arrigoni, Alberto et al. “Analysis RNA-seq and Noncoding RNA.” Methods in molecular biology (Clifton, N.J.) vol. 1480 (2016): 125-35. doi:10.1007/978-1-4939-6380-5_11
SECTIONS
TutorialsIn-house software
Blog
News
Find Jobs
History
Definition of Bioinformatics
ENCYCLOPEDIA
COVID-19
⚬ Timeline of outbreak
⚬ Blog
⚬ Scientific Facts
⚬ Daily statistics
⚬ News
TOOLS
1. SNP Tools
2. GWAS Tools
3. Genotype Imputation Tools
4. MSA tools
5. CNV tools
6. WGA tools
7. RNA-seq tools
8. Proteomics Tools
9. Nucleic Acid Structure Analysis Tools
10. Nucleic Acid Sequence Analysis Tools
11. Sequence Logo Tools
12. In-house Software Tools
Find thousands of Bioinformatics and Life Science software tools and databases in the newly launched
Ads
Ads
Ads