Bioinformatics software: a comprehensive guide
Learning objectives: Get an overall understanding of bioinformatics resources.
Introduction
Bioinformatics is a rapidly growing field that combines biology, computer science, and statistics to analyze and interpret large-scale biological data. With the advent of high-throughput sequencing technologies, there has been an explosion in the amount of biological data generated. This has led to the development of a wide range of bioinformatics software that can help researchers to analyze and interpret this data.
This comprehensive guide discusses the different types of bioinformatics software available and their applications.
Sequence alignment software

Sequence alignment is a fundamental tool in bioinformatics used to compare and identify similarities or differences between two or more biological sequences, such as DNA or protein sequences. This process is essential for understanding the structure and function of these sequences and predicting their evolution over time. Sequence alignment software is used for various purposes, including phylogenetic analysis, gene prediction, and functional annotation. In addition to these applications, sequence alignment is also used in drug discovery and development. It can help identify potential drug targets by comparing the sequences of proteins involved in disease pathways.
The sequence alignment process involves comparing one sequence's nucleotide or amino acid sequence to another. This comparison can reveal similarities and differences between the sequences, which can help identify important features such as conserved domains, motifs, and functional residues. Several methods exist for performing sequence alignment, including pairwise alignment, multiple sequence alignment, and profile alignment.
Pairwise alignment involves comparing two sequences and identifying the regions of similarity and difference. This method is useful for comparing two closely related sequences, such as those from different strains of the same species. Multiple sequence alignment, on the other hand, involves aligning three or more sequences to each other. This method is useful for comparing sequences from different species or different genes within the same species. Profile alignment is a more advanced method that involves aligning a sequence to a pre-existing alignment profile. This method is useful for identifying conserved domains and motifs within a set of related sequences.
Some commonly used sequence alignment software include Clustal Omega, MUSCLE, and MAFFT. Clustal Omega is an open-source multiple-sequence alignment tool widely used for DNA and protein sequences. It uses a fast algorithm and can handle large datasets with ease. Conversely, MUSCLE is a highly accurate tool that uses an iterative algorithm to refine the alignment. This makes it particularly useful for aligning closely related sequences. MAFFT is another popular tool that can handle various sequence types and sizes, making it suitable for various applications. It also has several advanced features, such as handling incomplete or fragmented sequences.
The accuracy of sequence alignment software is critical for the success of many bioinformatics applications. For example, accurate sequence alignment is essential for phylogenetic analysis, which is used to reconstruct the evolutionary history of species based on their genetic sequences. Similarly, accurate sequence alignment is essential for gene prediction, which involves identifying the location and function of genes within a genome. Inaccurate alignment can lead to incorrect predictions and hinder the progress of scientific research.As biological data is generated, the need for accurate and efficient sequence alignment software will only grow. Developing new and improved algorithms for sequence alignment is an ongoing area of research in bioinformatics. Researchers are working to develop algorithms that can handle larger datasets, align sequences with greater accuracy, and identify conserved domains and motifs more effectively. These advancements will help to unlock new insights into the natural world and improve our understanding of biological systems.
Browse sequence alignment software tools in Bioinformatics Software DB
Related tutorials
- Introduction to sequence comparison
- Pair-wise sequence alignment
- Pair-wise sequence alignment methods
- Construction of substitution matrices
- DNA Sequence Alignment How to
- How to select the right substitution matrix?
- Introduction to Information Theory and Its Applications to DNA and Protein Sequence Alignments
- Multiple sequence alignment (MSA) Methods
Genome annotation software
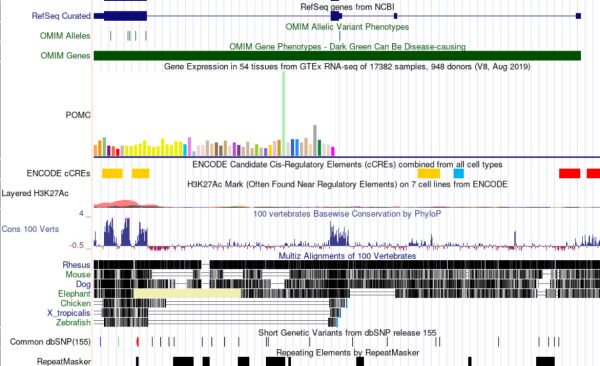
Genome annotation software has become a crucial tool in genomics, providing researchers with a more efficient and accurate means to annotate genomes. With the availability of high-throughput sequencing technologies, there has been an explosion in the amount of biological data generated. This has led to the need for genome annotation software that can help researchers accurately predict gene locations and their corresponding functions within a genome sequence.
Various software tools have been developed to address this need, and these tools use a combination of algorithms and methods to accurately predict gene locations and functions. Prokka, MAKER, and RAST are some of the most commonly used genome annotation software researchers rely on to annotate genomes accurately and efficiently.
Prokka is a popular tool designed to annotate bacterial and archaeal genomes. It uses a combination of software tools to predict genes, identify functional domains, and annotate the genome. Prokka is highly efficient and can produce high-quality annotations with minimal user intervention.
Conversely, MAKER is a more complex tool that can handle both prokaryotic and eukaryotic genomes. It utilizes evidence-based and ab initio methods to annotate the genome, providing a more comprehensive genome structure and function analysis. MAKER is highly customizable; users can configure the software to suit their needs and requirements.
RAST is a web-based tool that provides rapid annotation of microbial genomes, making it a popular choice for researchers who require a quick and easy solution for genome annotation. RAST uses a highly efficient annotation pipeline to process large datasets with minimal computational resources.
The use of genome annotation software has revolutionized the field of genomics by making it possible to annotate genomes accurately and efficiently. These software tools have enabled researchers to focus on other aspects of their research while the software handles the annotation process. Additionally, genome annotation software has led to significant advancements in our understanding of genetic diseases and their potential treatments.
The accuracy of genome annotation software is critical for accurate predictions of gene locations and functions. It is essential for understanding the structure and function of genes and their role in disease pathways. The continued development and improvement of genome annotation software will be critical in unlocking new insights into the natural world and developing new disease treatments.
See more genome annotation software topic in Bioinformatics Software DB - annotation
Protein structure prediction software
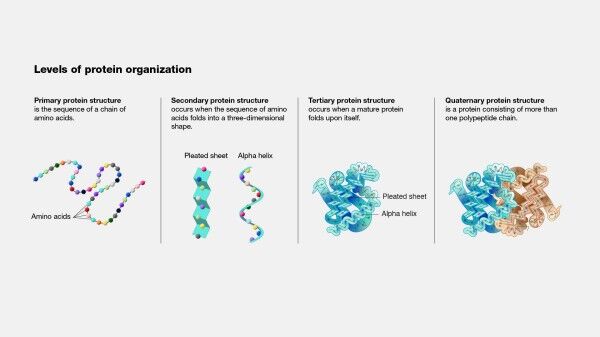
Protein structure prediction is a complex and challenging task that remains a significant area of research in biochemistry. The three-dimensional structure of a protein is essential for understanding its biological function and interactions with other molecules. The protein structure prediction process involves several steps: template selection, alignment, model building, refinement, and validation. These steps are critical in ensuring accurate protein structure prediction, essential for developing new disease treatments, and advancing our understanding of the natural world.
Template selection is the first step in the protein structure prediction process. It involves identifying proteins with similar sequences to the target protein and using these as templates to build a model. The model's accuracy relies heavily on the quality of the templates used. Therefore, researchers must select templates closely related to the target protein to ensure the model's accuracy.
Alignment, the second step of the protein structure prediction process, involves aligning the target protein sequence with the template sequences. Accurate alignment is essential for generating an accurate model of the protein structure. Researchers use various algorithms and methods to align the sequences, including pairwise, multiple-sequence, and profile alignment.
Model building, the third step in the protein structure prediction process, involves generating a protein structure model based on the alignment. Researchers use various algorithms and computational methods to build the model, including homology modeling, ab initio modeling, and hybrid methods. Homology modeling involves using the templates to build a model, while ab initio modeling involves building a model from scratch.
Refinement is the fourth step in the protein structure prediction process, and it involves optimizing the model to improve its accuracy. Researchers use various algorithms and methods to refine the model, including molecular dynamics simulations, energy minimization, and side-chain optimization.
Validation is the final step in the protein structure prediction process, and it involves evaluating the quality of the model generated. Researchers use various tools and methods to validate the model, including Ramachandran plots, MolProbity, and QMEAN.
Protein structure prediction tools have revolutionized how researchers predict the structure of proteins. By using a combination of algorithms and computational methods, tools like Phyre2, I-TASSER, and Rosetta have made it possible to generate highly accurate models of protein structures. Phyre2, for example, uses homology modeling and ab initio methods to predict the location of active sites, ligand-binding sites, and disordered regions. I-TASSER, on the other hand, uses threading and ab initio methods to generate models while providing information about the quality of the models generated. Rosetta is a more advanced tool that uses a Monte Carlo algorithm to generate models and can perform molecular dynamics simulations and predict the stability of the protein structure.
Despite the advances in protein structure prediction, predicting the structure of a protein accurately remains challenging. Accurate protein structure prediction relies on various factors, including the templates' quality, the alignment's accuracy, the model building's quality, and the refinement strategies employed. In addition, the accuracy of the prediction tools depends on the amount and quality of data used to train and validate them. Therefore, it is essential to continually improve the accuracy of the prediction tools and the quality of the data used to train them.
Protein structure prediction is an active area of research that has the potential to unlock new insights into the natural world and develop new treatments for diseases. In recent years, deep learning models have shown promise in improving the accuracy of protein structure prediction. These models use artificial neural networks to learn the complex relationships between amino acid sequences and protein structures. Researchers are also exploring new algorithms and computational methods to improve the accuracy of the prediction tools. These advancements will help unlock new insights into the natural world and improve our understanding of biological systems.
Single nucleotide polymorphism software
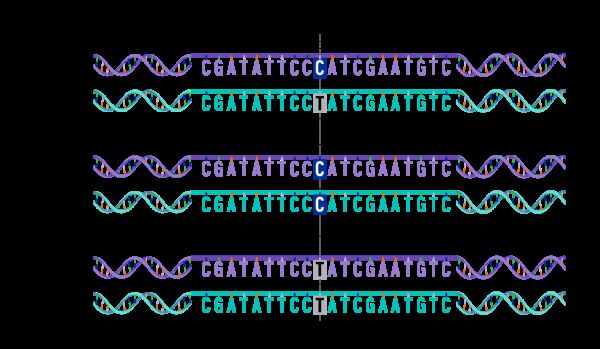
In genetics, single nucleotide polymorphisms (SNPs) have become a crucial aspect of genomic research. SNPs are the most common type of genetic variation that occurs when a single nucleotide (A, T, C, or G) in the genome differs between individuals. These variations play a significant role in the development of diseases, drug response, and even the evolution of species. Therefore, identifying and analyzing SNPs are essential for understanding the genetic basis of diseases and developing personalized treatments.
Fortunately, bioinformatics has developed efficient and accurate SNP software to identify and analyze SNPs in genomic data over the years. SNP software uses various algorithms and methods to identify SNPs accurately and efficiently, enabling researchers to analyze large-scale genomic data with ease and accuracy.
One of the widely used SNP software is PLINK. This powerful tool can easily handle large datasets and perform a broad range of analyses, including association studies, linkage disequilibrium analysis, and genome-wide association studies (GWAS). The software is highly flexible and customizable; researchers can configure it to suit their needs and requirements. PLINK has been cited in thousands of scientific publications and has become a go-to tool for researchers worldwide.
Another popular SNP software is GATK (Genome Analysis Toolkit), which provides a comprehensive SNP calling, variant discovery, and genotyping toolkit. GATK is highly accurate, robust, and widely used in clinical settings for genetic testing. The software is highly customizable; users can configure it to suit their needs and requirements.
SAMtools is a suite of tools for manipulating and analyzing high-throughput sequencing data. It includes various tools for SNP calling, variant detection, and genotyping. SAMtools is highly efficient and can easily handle large datasets, making it a valuable tool for bioinformatics researchers.
The accurate identification and analysis of SNPs are critical for developing personalized treatments and improving patient outcomes. Using SNP software, researchers can identify disease-causing SNPs and develop new treatments, paving the way for innovative solutions to real-world problems.
SNP software has revolutionized genetic research, making it easy to analyze large-scale genomic data easily and accurately. These tools have enabled researchers to identify disease-causing SNPs, develop new treatments, and improve patient outcomes. As the amount of biological data generated increases, the need for SNP software will only grow, providing exciting opportunities for future research and discovery.
More Single nucleotide polymorphism related software tools in Free Single nucleotide polymorphism (SNP) Analysis Tools - Software and Resources
Genome-wide Association Study (GWAS) Software

Genome-wide Association Study (GWAS) Software has transformed the field of genomics, providing researchers with an efficient and accurate means to identify genetic variations associated with diseases and other complex traits. The process involves scanning an individual's genome to identify single nucleotide polymorphisms (SNPs) associated with a particular trait or disease. These SNPs can then be used to develop diagnostic tools, therapies, and personalized treatments.
The ability to analyze large-scale genomic data has paved the way for exciting discoveries and innovative solutions to real-world problems. However, analyzing GWAS data requires advanced bioinformatics tools for complex statistical analyses and large-scale genomic data. This is where GWAS software comes into play!
PLINK, SNPTEST, and EIGENSOFT are some of the most commonly used GWAS software. PLINK is a powerful tool that can easily handle large datasets and perform a broad range of analyses, including association studies, linkage disequilibrium analysis, and genome-wide association studies (GWAS). The software is highly flexible and customizable; researchers can configure it to suit their needs and requirements. PLINK has been cited in thousands of scientific publications and has become a go-to tool for researchers worldwide.
SNPTEST provides various tools for GWAS data analysis, including quality control, imputation, and association testing. It is highly efficient and can easily handle large datasets, making it a valuable tool for bioinformatics researchers. The software is designed to be highly user-friendly, making it accessible to researchers of all levels of expertise.
EIGENSOFT provides various tools for population structure analysis, association testing, and principal component analysis. This highly efficient software can easily handle large datasets, making it a valuable tool for bioinformatics researchers. The highly customizable software allows researchers to configure it to suit their needs and requirements.
GWAS software has significantly advanced our understanding of genetic diseases and their potential treatments. The ability to analyze large-scale genomic data with ease and accuracy has paved the way for exciting discoveries and innovative solutions to real-world problems.
However, the accuracy of GWAS software is critical for accurate predictions of genetic variations associated with diseases and other complex traits. Inaccurate data analysis can lead to incorrect predictions, hindering scientific research's progress.
As biological data is generated, the need for accurate and efficient GWAS software will only grow. Researchers are developing algorithms that can handle larger datasets, identify genetic variations associated with diseases and other complex traits with greater accuracy, and provide researchers with a means to validate their findings more efficiently. These advancements will help unlock new insights into the natural world and improve our understanding of biological systems.
Check out Free Genome-wide Association Study (GWAS) Tools - Software and Resources
Copy number variation (CNV) analysis software
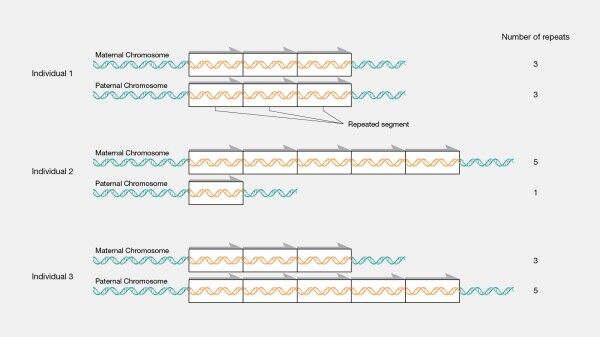
Copy number variations (CNVs) are a common form of genetic variation that refers to the gain or loss of genomic segments. These segments are usually longer than single nucleotides and range from a few hundred base pairs to several million. CNVs have been implicated in various diseases, including cancer, schizophrenia, and autism. Therefore, the accurate identification and analysis of CNVs are essential for understanding the genetic basis of these diseases and developing personalized treatments.
In recent years, there has been a significant increase in the amount of genomic data generated, leading to a growing need for efficient and accurate CNV analysis software. CNV analysis software has been developed to address this need, providing researchers with a powerful and efficient means to detect CNVs in genomic data. These software tools use various algorithms and methods to accurately identify CNVs in large-scale genomic data, enabling researchers to analyze and interpret the data easily and accurately.
CNV analysis software is a crucial tool in genomics, providing researchers with an efficient and accurate means to identify and analyze CNVs in genomic data. One of the most widely used CNV analysis software is CNVnator. This powerful tool can easily handle large datasets and perform various analyses, including CNV calling, genotyping, and visualization. This highly flexible and customizable software allows researchers to tailor it to their needs and requirements. It has been cited in numerous scientific publications and has become a go-to tool for researchers worldwide.
Another popular CNV analysis software is ExomeDepth, which provides a comprehensive CNV calling, genotyping, and visualization toolkit. ExomeDepth is highly accurate and robust, making it ideal for use in clinical settings for genetic testing. Like CNVnator, it is highly customizable, allowing users to configure the software to their specific needs and requirements.
VarScan is another suite of tools for detecting genomic variations, including CNVs, in high-throughput sequencing data. It includes various tools for CNV calling, visualization, and validation. VarScan is highly efficient and can easily handle large datasets, making it an invaluable tool for bioinformatics researchers.
CNV analysis software has revolutionized the field of genetic research, enabling researchers to analyze large-scale genomic data with ease and accuracy and facilitating the identification of disease-causing CNVs. CNV analysis software has significantly advanced our understanding of genetic diseases and their potential treatments. With the ability to analyze large-scale genomic data with ease and accuracy, these software tools have paved the way for exciting discoveries and innovations in genetics.
However, the accuracy of CNV analysis software is critical for accurate predictions of genetic variations associated with diseases and other complex traits. Inaccurate data analysis can lead to incorrect predictions, hindering scientific research's progress. Therefore, researchers are continually working to develop algorithms that are more accurate, efficient, and can handle larger datasets.
More copy number variation (CNV) software tools in Free Copy Number Variation (CNV) Analysis Tools - Software and Resources
Whole Genome Assembly (WGA) Software
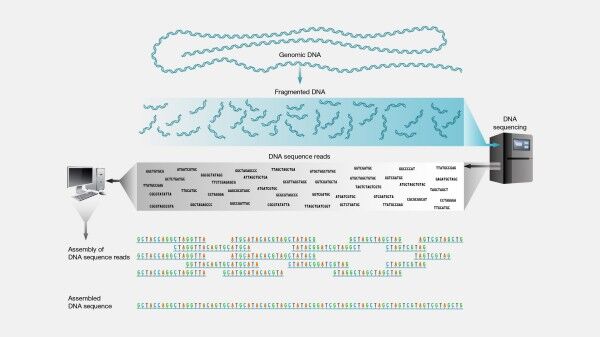
The whole genome assembly (WGA) software realm is fascinating. WGA software has revolutionized the field of genomics and has opened up a range of possibilities for researchers looking to understand the genetic makeup of organisms.
Assembling the entire genome sequence of an organism is a complex and challenging process, requiring specialized algorithms and computational methods to ensure accuracy. Over the years, various WGA software tools have been developed to address this need, providing researchers with various options.
WGA software has enabled researchers to comprehensively analyze an organism's genetic makeup, making it an essential tool in genomics. With the increasing amount of biological data generated daily, the need for accurate and efficient WGA software will only grow, providing exciting opportunities for future research and discovery.
SPAdes, ABySS, and Canu are some of the most commonly used WGA software that has transformed the field of genomics. SPAdes is a highly flexible and customizable tool that can easily handle large datasets and perform various genome assembly tasks for small bacterial genomes to large eukaryotic genomes. It combines de Bruijn graph construction, paired-end reads, and hybrid approaches to assemble highly accurate and contiguous genomes.
ABySS, on the other hand, provides various tools for genome assembly, including de Bruijn graph construction and assembly, error correction, and scaffolding. It is highly efficient and can easily handle large datasets, making it an invaluable asset for genomics researchers. ABySS uses a range of strategies to improve genome assembly quality, including using mate-pair and paired-end reads and incorporating additional data sources.
Canu, a more advanced WGA software, is specifically designed for long-read sequencing technologies, such as PacBio and Oxford Nanopore. This highly accurate and robust tool enables researchers to easily assemble high-quality genome sequences using error correction, trimming, and scaffolding. Canu can easily handle large datasets, making it a valuable tool for researchers with large and complex genomes.
WGA software has paved the way for exciting discoveries and innovations in genetics, providing researchers with a comprehensive view of an organism's genetic makeup. However, the accuracy of WGA software is critical for accurate predictions of an organism's genetic makeup, which is essential for developing new treatments for diseases and advancing our understanding of biological systems.
Therefore, researchers are continually working to develop algorithms that are more accurate, efficient, and can handle larger datasets. With the increasing amount of biological data generated daily, the need for accurate and efficient WGA software will only grow, providing exciting opportunities for future research and discovery.
WGA software will continue to be crucial in advancing our understanding of biological systems and developing new disease treatments. The need for accurate and efficient WGA software will only grow, providing exciting opportunities for researchers to unlock new insights into the natural world and develop innovative solutions for real-world problems.
In the future, WGA software will be key to unlocking discoveries and innovations in genetics. With continued development and improvement, this powerful tool will continue to pave the way for exciting genetic breakthroughs, enabling researchers to develop new treatments for diseases, improve patient outcomes, and revolutionize how we approach to disease diagnosis and treatment.
WGA software has transformed the field of genomics, enabling researchers to identify new genes, understand their functions, and predict their interactions with other genes. The accuracy of WGA software is critical for accurate predictions of an organism's genetic makeup, which is essential for developing new treatments for diseases and advancing our understanding of biological systems.
Therefore, WGA software's continued development and improvement will enable researchers to unlock new insights into the natural world and develop innovative solutions to real-world problems. With increasing biological data generated daily, WGA software will be critical in advancing our understanding of biological systems and developing new disease treatments.
See also Free Whole Genome Assembly (WGA) Analysis Tools - Software and Resources
Related tutorials
- The Principles of WGS Sequencing and Automated Fragment Assembly
- Introduction to sequence assembly
- Genome assembly Quality Metrics
- Sequence Assembly Practical
RNA-seq Software
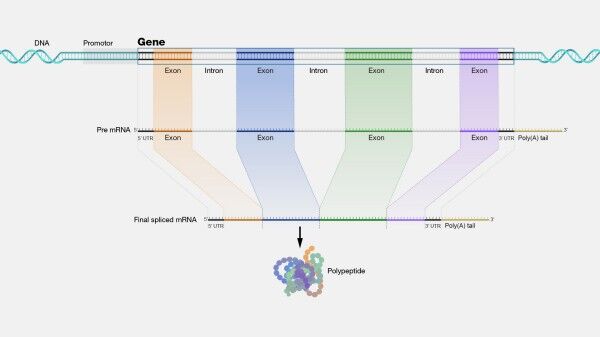
RNA sequencing (RNA-seq) has revolutionized the field of genomics and has become one of the most widely used methods for studying gene expression, alternative splicing, and transcriptome analysis. RNA-seq technology has enabled researchers to detect and quantify RNA molecules in a sample, providing a comprehensive view of the transcriptome of an organism.
RNA-seq software has been developed to address the need for efficient and accurate RNA-seq data analysis. These software tools use various algorithms and methods to identify differentially expressed genes, splice variants, and novel transcripts, enabling researchers to analyze and interpret RNA-seq data easily and accurately.
One of the most popular RNA-seq software is STAR (Spliced Transcripts Alignment to a Reference), a high-performance RNA-seq aligner that can easily handle large datasets. STAR is highly accurate and efficient, enabling researchers to analyze RNA-seq data with unparalleled sensitivity and specificity. STAR can identify novel splice junctions, making it a valuable tool for discovering new transcripts and splice variants.
Another widely used RNA-seq software is HISAT2, a fast and sensitive RNA-seq aligner that can easily handle large datasets. HISAT2 is highly accurate and efficient, enabling researchers to analyze RNA-seq data with high sensitivity and specificity. HISAT2 can identify novel splice junctions and transcripts, making it a valuable tool for discovering new transcripts and splice variants.
RSEM (RNA-Seq by Expectation-Maximization) is another widely used RNA-seq software that can accurately estimate gene and isoform expression levels. RSEM is highly accurate and efficient, enabling researchers to analyze RNA-seq data with high sensitivity and specificity. RSEM can easily handle large datasets, making it a valuable tool for analyzing RNA-seq data from various organisms.
RNA-seq software has significantly improved our understanding of gene expression and regulation. By identifying novel transcripts and splice variants, we can deepen our understanding of the structure and function of genes and their role in disease pathways. The accurate analysis of RNA-seq data can help identify differentially expressed genes, providing insights into the function of genes in different biological processes. This is essential for developing new disease treatments and advancing our understanding of biological systems.
However, the accuracy of RNA-seq software is critical for accurate predictions of differential gene expression and splice variants, which is essential for understanding the structure and function of genes and their role in disease pathways. Inaccurate data analysis can lead to incorrect predictions, hindering scientific research's progress. Therefore, researchers are continually working to develop algorithms that are more accurate, efficient, and can handle larger datasets.
As the amount of biological data generated daily increases, the need for accurate and efficient RNA-seq software will only grow, providing exciting opportunities for future research and discovery. RNA-seq software has the potential to revolutionize the way we approach disease diagnosis and treatment. With continued development and improvement, this powerful tool will pave the way for exciting breakthroughs in genomics, enabling researchers to develop new treatments for diseases, improve patient outcomes, and unlock new insights into biological systems.
RNA-seq software has paved the way for exciting discoveries and innovations in genomics. The accurate RNA-seq data analysis has enabled researchers to identify genes linked to specific diseases, paving the way for targeted treatments. RNA-seq software has also enabled researchers to study alternative splicing, an essential mechanism for generating protein diversity. The accurate analysis of alternative splicing can help identify novel transcripts, providing insights into the structure and function of genes.
Moreover, RNA-seq software has opened up a new range of possibilities for researchers studying non-coding RNA. Non-coding RNA is essential in regulating gene expression and has been linked to various diseases. RNA-seq software can study the expression of non-coding RNA, providing insights into their function and role in biological systems.
With the increasing amount of biological data generated daily, the need for accurate and efficient RNA-seq software will only grow, providing exciting opportunities for future research and discovery. RNA-seq software holds immense potential to revolutionize the field of genomics, enabling researchers to develop new treatments for diseases, improve patient outcomes, and unlock new insights into biological systems.
Numerous RNA-seq software tools in Free RNA-seq Analysis Tools - Software and Resources
Nucleic Acid Structure Analysis Software
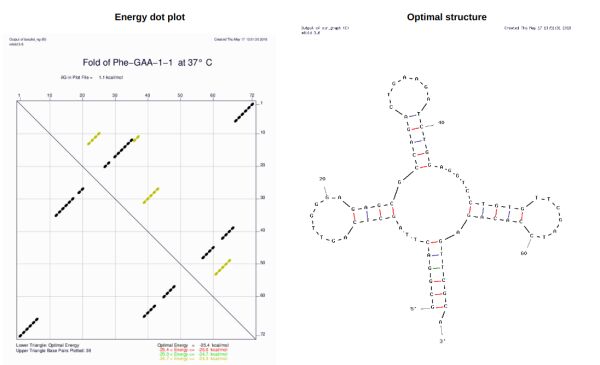
Nucleic acid structure analysis software - an effective tool that can analyze and interpret the structure of nucleic acids, such as DNA and RNA, with unparalleled accuracy. The structure of nucleic acids is a fundamental component of understanding their function and interactions with other molecules. It is essential to unlocking the mysteries of the biological world.
The development of nucleic acid structure analysis software has opened up a new range of possibilities for researchers studying the structure and function of nucleic acids. The software provides a time-efficient, comprehensive and accurate means to analyze and interpret the structure of nucleic acids. The accurate analysis of nucleic acid structure can help identify novel structural motifs and interactions, providing insights into the structure and function of nucleic acids.
Another widely used nucleic acid structure analysis software is 3DNA, a comprehensive toolkit for analyzing the structure of nucleic acids. 3DNA provides a range of tools for analyzing the three-dimensional structure of nucleic acids, including base-pair and backbone parameters, helix geometry, and molecular graphics. Its efficient algorithms can easily handle large datasets, making it an invaluable asset for genomics researchers.
RNAstructure is another software that uses various algorithms and methods to identify structural motifs and interactions. Its highly efficient and accurate predictions make it an essential tool for understanding the function of RNA molecules, providing insights into the structure and function of nucleic acids. RNAstructure is highly efficient and can easily handle large datasets, making it an invaluable asset for RNA researchers.
Using nucleic acid structure analysis software has significantly advanced our understanding of nucleic acid structure and function. With accurate predictions, we can deepen our understanding of the structure and function of genes, paving the way for exciting discoveries in genomics.
However, the accuracy of nucleic acid structure analysis software is critical for understanding the structure and function of nucleic acids and their role in disease pathways. Inaccurate data analysis can lead to incorrect predictions, hindering scientific research's progress.
As the amount of biological data generated daily increases, the need for accurate and efficient nucleic acid structure analysis software will only grow, providing exciting opportunities for future research and discovery. The future of nucleic acid structure analysis software looks bright, holding the immense potential to transform the field of genomics and revolutionize how we approach to disease diagnosis and treatment.
Nucleic acid structure analysis software - a tool that unlocks the secrets of the biological world, revealing the intricacies of nucleic acids with unparalleled accuracy. The possibilities for future discoveries are endless, and with continued development and improvement, this powerful tool will pave the way for exciting breakthroughs in genomics.
Nucleic acid structure analysis software is the tool that has allowed researchers to explore the depths of the genetic code, enabling scientists to examine and interpret the intricacies of DNA and RNA. This software is a critical component of modern-day bioinformatics and is helping to unlock the secrets of the biological world. The possibilities for future discoveries are endless, and with continued development and improvement, this powerful tool will pave the way for exciting breakthroughs in genomics.
For additional software, see Nucleic Acid Structure Analysis Tools - Software and Resources
DNA Sequence Analysis Software
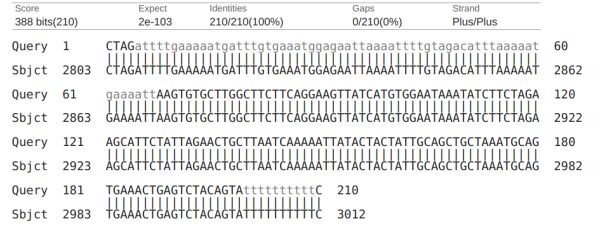
DNA Sequence Analysis Software is a crucial tool in genomics, enabling scientists to unlock the secrets of the genetic code and develop new disease treatments. This powerful and efficient technology has revolutionized the field of genomics, providing researchers with a means to analyze and interpret DNA sequences accurately and efficiently.
The accuracy of DNA sequence analysis software is essential for understanding the structure and function of genes and their role in disease pathways. The most commonly used DNA sequence analysis software tools, such as BLAST, Bowtie, and BWA, have transformed the field of genomics research. These incredible algorithms can easily handle large datasets, allowing researchers to predict an organism's genetic makeup accurately.
BLAST (Basic Local Alignment Search Tool) can compare a query sequence to a database of sequences with high accuracy and sensitivity. Bowtie can align millions of short DNA sequences to a reference genome with high accuracy and speed. BWA (Burrows-Wheeler Aligner) can align short DNA sequences with high sensitivity and specificity to a reference genome. This software is at the forefront of the genomics field, allowing researchers to analyze vast amounts of data in a fraction of the time it would take manually.
However, the power of DNA Sequence Analysis Software doesn't end with accuracy alone. The speed and efficiency of this software are also crucial in handling the increasing amount of biological data being generated daily. The field of genomics is advancing rapidly, and the need for DNA sequence analysis software that can handle larger datasets will become more pressing, providing exciting opportunities for future research and discovery.
Researchers are continually working to develop algorithms that are more accurate, efficient, and can handle larger datasets. As genomics advances, the need for DNA sequence analysis software to handle increasing biological data will become more pressing. This provides exciting opportunities for future research and discovery.
DNA Sequence Analysis Software has not only paved the way for exciting discoveries and innovations in genetics but has also enabled researchers to better understand the complex biological systems that govern life. With the increasing amount of biological data being generated daily, the need for accurate and efficient DNA Sequence Analysis Software will only grow, providing researchers with the ability to develop new treatments for diseases, improve patient outcomes, and unlock new insights into biological systems.
Genome annotation is another crucial task in bioinformatics that uses sequence alignment and analysis software tools (see below). It involves the identification of gene locations and their corresponding functions within a genome sequence, which is essential for understanding the structure and function of genes and their role in disease pathways. Genome annotation software has been developed to automate and simplify genome annotation. These tools utilize algorithms and methods to accurately predict gene locations and functions. The most commonly used genome annotation software include Prokka, MAKER, and RAST.
The accuracy of DNA Sequence Analysis Software is critical for understanding the structure and function of genes and their role in disease pathways. DNA sequence analysis software is used for various purposes, including gene prediction, functional annotation, and phylogenetic analysis. Sequence alignment software is used for various purposes, including phylogenetic analysis, gene prediction, and functional annotation.
See also DNA Sequence Analysis Tools - Software and Resources
Phylogenetics Software

The history of life on Earth is a complex tapestry of events over millions of years. From the tiniest microbes to the largest mammals, each species has its unique story to tell. For centuries, scientists have been piecing together the puzzle of life's origins, trying to unravel the mysteries of our biological past. One of the most critical developments in this field has been the advent of phylogenetics software.
Phylogenetics software has revolutionized the field of evolutionary biology, enabling researchers to study the evolutionary relationships between different species with unparalleled accuracy and efficiency. These tools have led to significant advancements in our understanding of the history of life on Earth, providing insights into the origins and diversification of different species.
Phylogenetics software utilizes algorithms and computational methods to analyze DNA and protein sequences, identify their similarities and differences, and construct evolutionary trees that illustrate the relationships between species. By comparing the genetic makeup of different organisms, researchers can reconstruct the history of life on Earth, tracing the origins of different species and identifying the mechanisms of evolution that have shaped our planet.
One of the oldest and most widely used phylogenetics software is PHYLIP, developed in the 1980s. PHYLIP provides a range of tools for phylogenetic analysis, including distance-based, maximum likelihood, and Bayesian methods. The software is highly efficient and can easily handle large datasets, making it an invaluable asset for evolutionary biologists.
Another popular phylogenetics software is MrBayes, a Bayesian inference program that uses Markov chain Monte Carlo (MCMC) methods to construct evolutionary trees. MrBayes is highly accurate and efficient, making it an essential tool for evolutionary biologists. It can easily handle large datasets and provides various tools for analyzing and visualizing phylogenetic trees.
BEAST (Bayesian Evolutionary Analysis Sampling Trees) is another widely used phylogenetics software. This software employs Bayesian methods to construct phylogenetic trees and estimate evolutionary parameters, such as divergence times and substitution rates. BEAST is highly efficient and accurate, making it an essential tool for evolutionary biologists working on large and complex datasets.
Phylogenetics software has also led to identifying new species and discovering previously unknown relationships between species. By identifying these relationships, researchers can better understand the mechanisms of evolution, providing insights into the factors that drive the diversification of life on Earth.
Moreover, phylogenetics software has revolutionized the field of conservation biology. Scientists use these tools to study the evolutionary relationships between different species and identify the most at risk of extinction. This knowledge is used to develop conservation strategies that protect endangered species and preserve biodiversity.
The use of phylogenetics software has also led to significant advancements in medicine. By studying the evolutionary relationships between different species, researchers can identify the genetic basis of diseases and develop new treatments. For example, scientists have used phylogenetics software to study the evolutionary history of HIV and develop new treatments that target specific virus strains.
As we continue to uncover the secrets of our biological past, phylogenetics software will play an increasingly important role in our understanding of the natural world. By studying the evolutionary relationships between species, we can gain insights into the complex interplay between genetics, ecology, and environmental factors. This knowledge can be applied to various fields, from conservation biology to medicine, providing us with the tools to create a brighter future for our planet and its inhabitants.
More Free Phylogenetics Tools - Software and Resources
Related tutorials
Chip-seq Analysis Software: A Revolution in Genomics Research
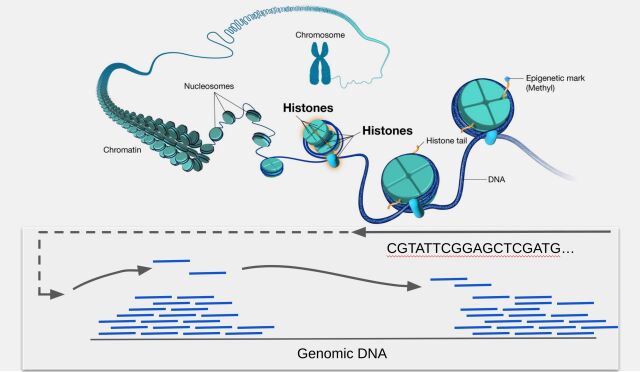
In modern genomics research, the Chromatin immunoprecipitation sequencing (ChIP-seq) technique has revolutionized how researchers study the interactions between proteins and DNA at a genome-wide scale. As a result, researchers can identify transcription factor binding sites, histone modifications, and other regulatory elements.
However, analyzing ChIP-seq data is a complex and challenging task requiring specialized software tools to accurately process and interpret the data. This is where Chip-seq analysis software comes in. It provides various tools for analyzing ChIP-seq data, including peak calling, peak annotation, and differential binding analysis. These tools enable researchers to identify regulatory elements, identify transcription factor binding sites, and study the interactions between proteins and DNA.
A popular Chip-seq analysis software is SICER (Spatial Clustering for Identification of ChIP-Enriched Regions). It is a highly accurate and efficient tool that uses a clustering algorithm to identify ChIP-seq peaks. It also provides a range of tools for downstream analysis, including peak annotation, differential binding analysis, and motif discovery. SICER is also highly customizable, allowing users to adjust the parameters to suit their needs.
The widespread use of Chip-seq analysis software has significantly advanced our understanding of gene regulation and epigenetic modifications. Researchers can gain insights into gene expression mechanisms and epigenetic modifications' role in disease pathways by identifying transcription factor binding sites and other regulatory elements. This has opened the door to exciting discoveries in genomics.
However, analyzing ChIP-seq data is still challenging, and researchers are continually developing new algorithms and methods to improve the accuracy and efficiency of Chip-seq analysis software. With the increasing amount of biological data generated daily, the need for accurate and efficient Chip-seq analysis software will only grow, providing exciting opportunities for future research and discovery.
As we uncover the secrets of our biological past, developing cutting-edge Chip-seq analysis software becomes even more essential. The ChIP-seq data analysis is crucial to understanding gene expression and epigenetic modifications. Effective analysis software is necessary to process this technique's vast data. With innovative algorithms and improved analyses, Chip-seq analysis software will significantly advance our understanding of biological systems, paving the way for exciting discoveries and innovative solutions to real-world problems.
Researchers are continually working to improve Chip-seq analysis software, developing new methods and algorithms to handle the vast amounts of data generated and improve the analyses' accuracy and efficiency. From peak calling and peak annotation to differential binding analysis and motif discovery, Chip-seq analysis software has become an essential tool in genomics, paving the way for exciting discoveries and innovations.
As we unravel the natural world's mysteries, Chip-seq analysis software will play an increasingly important role in understanding the complex interplay between genetics, epigenetics, and environmental factors. With the help of Chip-seq analysis software, researchers can gain insights into gene expression mechanisms and epigenetic modifications' role in disease pathways. This knowledge can be applied to various fields, from conservation biology to medicine, providing us with the tools to create a brighter future for our planet and its inhabitants.
For more ChIP-seq software, see a collection in Bioinformatics Software DB
Metagenomics Software
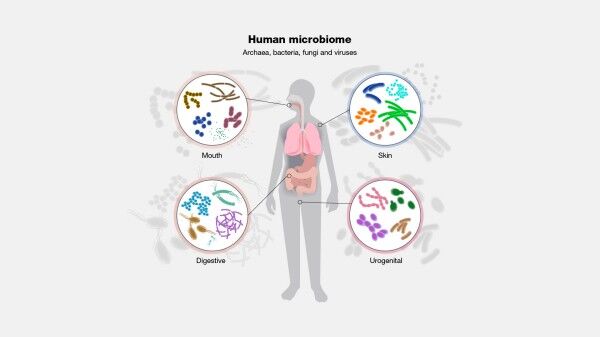
The study of genetic material recovered directly from environmental samples, known as metagenomics, has become a vital tool in genomics, enabling researchers to study the microbial communities' genetic makeup and provide insights into the complex interactions between microorganisms. Metagenomics refers to the study of the genetic material of microbial communities recovered from environmental samples such as soil, water, and air. It has transformed our understanding of microbial ecology and the role of microorganisms in various ecosystems.
At the forefront of this exciting field the cutting-edge tools and software known as metagenomics software. These software programs provide a range of powerful tools for analyzing metagenomic data, including sequence assembly, gene annotation, and taxonomic classification. With their help, researchers can identify the presence and abundance of different microorganisms, unlocking new insights into their complex interactions.
One of the most widely used metagenomics software is QIIME (Quantitative Insights Into Microbial Ecology). Its highly efficient and easily handles large datasets, making it an invaluable asset for metagenomics researchers. It provides various tools for analyzing metagenomic data, including sequence quality control, sequence alignment, and taxonomic classification.
MG-RAST (Metagenomics Rapid Annotation using Subsystem Technology) is another metagenomics software making waves. Its powerful tools for analyzing metagenomic data, including sequence quality control, sequence assembly, and gene annotation, make it an essential tool for metagenomics researchers. It can also handle large datasets, making it a popular choice for studying microbial communities.
MetaPhlAn (Metagenomic Phylogenetic Analysis) employs a unique marker-based approach to identify microbial taxa present in a metagenomic sample, providing highly accurate and efficient taxonomic classification. This makes it an excellent choice for researchers looking to profile and estimate the abundance of microbial communities using shotgun metagenomic sequencing data.
Check out more Metagenomics software tools
Primer Design Software: A Key Tool in Molecular Biology Research
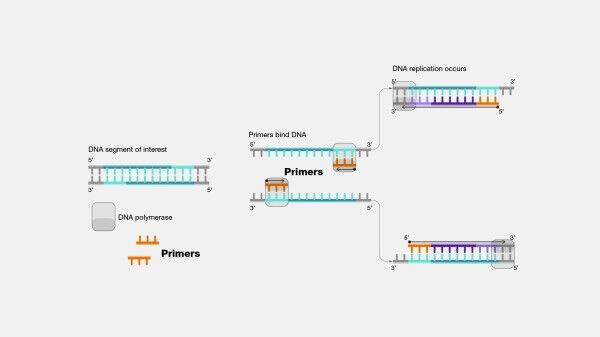
The Polymerase Chain Reaction (PCR) is a vital tool in molecular biology research, enabling researchers to amplify small amounts of DNA for further analysis. However, designing specific and efficient primers for PCR can be challenging, requiring specialized software tools to accurately design and optimize primers for specific targets. This is where Primer Design Software comes in.
Primer Design Software is a must-have tool for researchers looking to amplify target sequences with high accuracy and efficiency. This software utilizes various powerful tools, including sequence analysis, melting temperature calculations, and specificity checks, to help researchers design and optimize PCR primers. With the resulting specific and efficient primers, researchers can conduct further analysis and unlock exciting discoveries in bioinformatics. Don't miss the opportunity to take your research to the next level with Primer Design Software!
One of the most widely used primer design software is Primer3. It provides various tools for designing and optimizing PCR primers, including sequence analysis tools, melting temperature calculations, and specificity checks. Primer3 is highly efficient and can design primers for various applications, including genotyping, mutation detection, and quantitative PCR.
Another popular primer design software is OligoCalc. It provides various tools for designing and optimizing PCR primers, including melting temperature calculations and specificity checks. OligoCalc is highly efficient and can design primers for various applications, including quantitative PCR, mutagenesis, and gene cloning.
The accuracy of Primer Design Software is critical for understanding the structure and function of genes and their role in disease pathways. Accurate primer design enables researchers to amplify target sequences with high accuracy and efficiency, enabling them to carry out further analysis. Therefore, researchers are continually working to develop algorithms that are more accurate, efficient, and can handle larger datasets.
Like other bioinformatics software, Primer Design Software has significantly advanced our understanding of molecular biology, providing insights into the structure and function of genes and their role in disease pathways. Researchers can conduct further analysis by designing specific and efficient primers, from genotyping and mutation detection to quantitative PCR and gene expression analysis. With continued development and improvement, bioinformatics software will continue to pave the way for exciting breakthroughs in molecular biology, enabling researchers to develop new treatments for diseases, improve patient outcomes, and unlock new insights into biological systems.
For example, scientists now use artificial intelligence and machine learning algorithms to develop more accurate primer design software. These algorithms use large datasets to train machine learning models to predict the most efficient primers for specific targets, enabling more efficient and accurate primer design.
Primer Design Software has expanded beyond PCR amplification by developing new techniques such as CRISPR-Cas9 gene editing. Primer Design Software is now used to design the gRNA (guide RNA) sequences that direct the Cas9 protein to the target gene for editing. Accurate gRNA design is essential for precise and efficient gene editing, making Primer Design Software a crucial tool in gene editing research.
Find additional Primer design software
Systems Biology Software
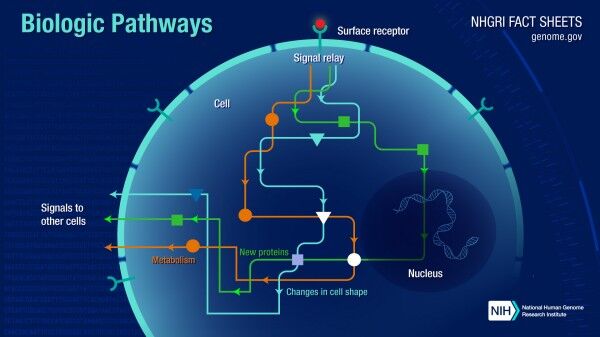
Systems biology software has revolutionized how researchers study complex biological systems by enabling them to model and analyze them as a whole rather than study each component in isolation. This approach has led to significant advancements in our understanding of biological systems, providing insights into the complex interplay between genetics, ecology, and environmental factors.
The development of systems biology software has been a breakthrough in biology. It has allowed researchers to simulate and model biological systems as a whole, providing a comprehensive view of the dynamics of biological systems. This approach has led to discoveries and insights into the complex interplay between genetics, environment, and other factors, potentially transforming our understanding of biological systems and improving human health and well-being.
Systems biology software utilizes mathematical and computational models to simulate the behavior of biological systems. These models integrate data from various sources, including genomics, transcriptomics, and proteomics, to provide a comprehensive view of biological systems' dynamics. By studying these models, researchers can identify the underlying mechanisms that govern biological systems and provide insights into how they function, how they can be manipulated to improve human health and well-being, and how they evolve over time.
MATLAB is one of the most widely used in systems biology, a programming language and software environment for numerical computing and data analysis. MATLAB provides various tools for modeling and simulating biological systems, including differential equation solvers, optimization algorithms, and statistical analysis tools. The software also has a user-friendly interface that allows researchers to easily analyze and visualize complex data.
Another popular systems biology software is BioNetGen, a rule-based modeling language for simulating complex biochemical systems. BioNetGen provides various tools for modeling and simulating biological systems, including stochastic and deterministic simulation methods. The software is widely used in systems biology and has been instrumental in advancing our understanding of biological systems' behavior.
COPASI (Complex Pathway Simulator) is another widely used systems biology software that provides tools for modeling and simulating biochemical networks. The software utilizes a range of mathematical models, including ordinary differential equations, stochastic simulation, and partial differential equations, to simulate the behavior of biological systems. COPASI also provides various tools for analyzing and visualizing simulation results, making it an essential tool in systems biology research.
Using systems biology software has significantly advanced our understanding of biological systems, providing insights into the complex interplay between genetics, ecology, and environmental factors. By studying biological systems, researchers can identify the underlying mechanisms that govern them, providing insights into how they function and can be manipulated to improve human health and well-being.
Systems biology software has also led to significant advancements in synthetic biology, providing researchers with the tools to engineer biological systems for specific purposes. By designing and simulating biological systems, researchers can identify the optimal conditions for these systems to function, enabling the development of new treatments for diseases and the production of biofuels and other useful products.
Explore more systems biology software tools in our database.
Data visualization software
Bioinformatics software has revolutionized modern scientific research, providing a means to analyze and interpret large-scale biological data more efficiently than ever. By combining biology, computer science, and statistics, bioinformatics software has made it possible to develop algorithms and software that can store, analyze, and visualize biological data, unlocking a vast range of possibilities for scientific discovery.
Data visualization is one of the most vital aspects of bioinformatics software. With the help of data visualization tools, researchers can explore and analyze complex biological data by presenting the data in an easily digestible visual format. These tools enable researchers to identify patterns and trends in data more efficiently, communicate research findings to others, and make data-driven decisions with greater confidence. Some of bioinformatics's most commonly used data visualization software includes R, Python, and Cytoscape.
R is a programming language and software environment widely used in bioinformatics. It provides various data analysis, manipulation, and visualization tools. The software is open-source, which means that it is free to use and can be modified by users to suit their needs. R has a vast user community, which makes it easy for new users to get started and learn the language.
Python is another programming language widely used in bioinformatics. It is known for its ease of use and versatility. Python provides a range of modules and packages for data analysis and visualization, making it a useful tool for researchers. Furthermore, Python is open-source, meaning it is free to use and can be modified by users to suit their needs.
Cytoscape is a popular tool for visualizing and analyzing biological networks, such as protein-protein interaction networks and metabolic pathways. The software provides a user-friendly interface that allows researchers to interact with network data and visualize it in various ways. Cytoscape has a range of plugins that can extend its functionality, making it a valuable tool for bioinformatics researchers.
In conclusion, bioinformatics has been transformed by the advent of bioinformatics software, which has provided researchers with the tools they need to analyze and interpret large-scale biological data more efficiently than ever before. With the increasing amount of biological data generated daily, bioinformatics software will continue to play a crucial role in advancing our understanding of biological systems and developing new disease treatments. Bioinformatics software's continued development and improvement will enable researchers to unlock new insights into the natural world and develop innovative solutions to real-world problems.
Biology includes many sub-disciplines, and there are always specialized bioinformatics software tools for each. You can browse bioinformatics software by topic in our Bioinformatics Software Database.
Related tutorials
- Genotype Imputation
- Best bioinformatics tools for beginners
- Applications of bioinformatics in various fields
