GRN Kernels, Character Identity Networks (ChIN), and Deep Homology
The kernel concept
The development of embryos in different species seems to follow similar patterns. For example, we can observe that human embryos have a tail around 30-35 weeks into the pregnancy (Figure 12).
Many species are visually close to indistinguishable from each other in the early stages of development (Video 1).
The observed similarities in embryo development have led to different various hypotheses and theories. One notable approach uses the kernel concept. This concept treats genomes as a blueprint for embryo development.
Video 1. Embryonic development - embryogenesis
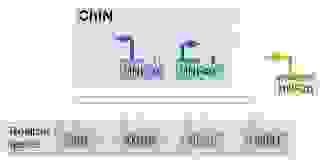
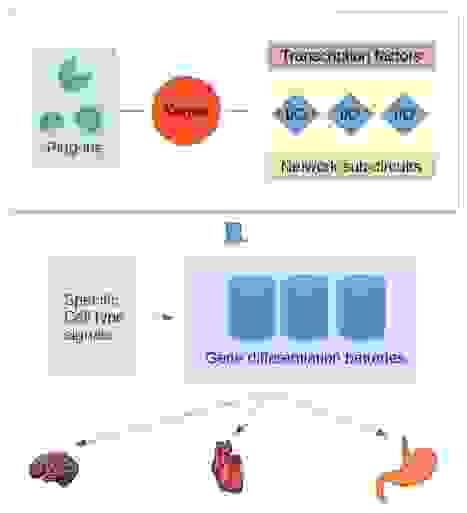
The kernel concept uses four separate gene regulatory components: 1. The kernel, 2. plug-ins, 3. input-output (I/O) switches, and 4. differentiation gene batteries (Figure 13).
Kernels are the core regulatory subunits that define the central fundamental developmental patterns. They are resistant to changes and rewiring and are therefore evolutionarily strongly conserved.
The strongly conserved kernels may be responsible for the stable animal body plans that we can track back over 500 million years.
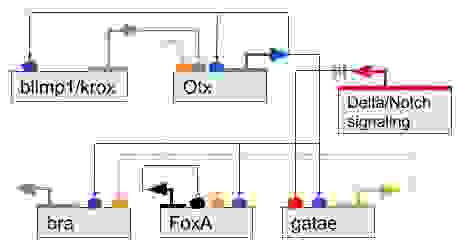
In Figure 14, we can see an example of a kernel that regulates the development of endomesoderm and is conserved in both Sea urchins and Starfishes.


Plug-ins are similarly, as kernels, small structurally conserved regulatory sub-circuitries. However, they do not specify the development of body parts.
Plug-ins are insertable modules into various gene regulatory schemes and, for example, affecting a limited set of transcription factors.
Input/output (I/O) switches regulate various other sub-networks and are the subject of evolutionary changes in the embryonal development processes. In many cases, the changes can be trivial, such as the size of body parts.
The determination of a body part size may only require a specific way to connect a body part patterning gene with cell cycle genes. A few examples are Drosophila haltere wing patterning, hox gene specifying the vertebrate morphology, and the fore-wing vs. hind-wing patterning.
Differentiation gene batteries are the executive branch consisting of specific groups of protein-coding genes. The gene batteries produce distinct proteins needed by each cell type and do not regulate any other genes or exercise other controls.
They are active at the end of the developmental processes and controlled by receiving cell-type-specific signals. The expression of specific differentiation gene batteries is a terminal process of differentiation.
The differentiation gene batteries may evolve by changes in their coding sequences, loosing, and gaining genes.
The character identity network (ChIN) concept
Before we dive into the character identity network, we need to define two things: 1. Character identity, and 2. character state. It is essential to understand the difference between these because they represent two separate evolutionary relationships.
While the character identity remains the same, it can appear in different forms as character states. Butterflies appendices have developed into four wings used for flying (Figure 15).
In cranefly, the other pair of appendices have developed into halteres, that insects use to sense the body position during a flight (Figure 16). Beetles' one of the appendix pairs have developed into wing covers, elytra (Figure 16).
An organ with one character identity can appear in different architectures, i.e., character states, as in the cases of the appendices of butterflies, craneflies, and beetles.


A core gene regulatory networks cause a developmental individualization of different body parts by utilizing a variety of effector genes.
These core gene regulatory networks are character identity networks (ChINs) that preserve the identity of a character but allow a set of effector genes to produce a variety of character states. Different realizations, i.e., different phenotypes of the same body part.
Character identity networks, as the name says, preserve the identity of an organ or a body part. This way, the hind wings of the butterfly (Figure 15) and the halteres of the cranefly (Figure 16) are homologous because the origin of each individualized body part is the same in both species (Figure 18).
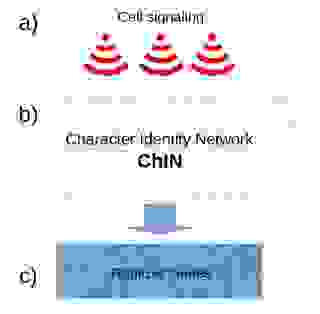
The ChIN concept has three organizational levels: 1. positional information through cell-cell signaling, 2. ChINs, and 3. realizer genes (Figure 18.)
1. Cell-cell signaling gives positional information to ChINs and activates them at specific locations. The signaling genes can be variable between species (Figure 18a).
2. The ChIN networks, activated by the cell-cell signaling, are well conserved in various species. The ChINs consist mostly of transcription factors, transcription co-factors, and long non-coding RNAs. ChINs specify the character identity and can regulate different realizer genes as target genes in various species, but do not necessarily determine the set of realizer genes (Figure 18b).
3. The realizer genes are genes that produce proteins and enzymes for a specific cell type, the workforce of the cells. This set of genes bear similarity to the differentiation gene batteries in the kernel concept. The disassociation of ChINs from the realizer genes explains how the phenotype or a character state can be a separate entity from the character identity (Figure 18c).
In Figure 19, is a real-world example of a ChIN of hepatocytes. Notice the positive feedback loop, which is characteristic in all ChINs. For the ChINs to work, all the transcription factors within a ChIN need to co-operate.
The necessity of the transcription factor co-operation may also be a reason for them to be well conserved. A single change in one of the factors is likely to damage the co-operation; Thus, keeping the ChINs conserved and preserving the character identity.
Charles Darwin well explained how a multitude of structures in various organisms arises through descent with modifications. However, how unique structures and phenotypes arise has been under debate for a long time. The character identity network (ChIN) concept could explain the rise of unique structures by an introduction of unique ChINs.
The Deep Homology Concept
Shubin, Tabin, and Carroll coined the term deep homology in 1997. Deep homology term covers conserved gene networks in the developmental differentiation processes in separate species.
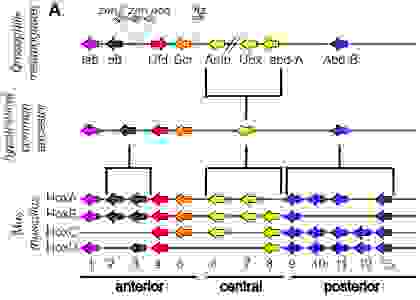
We can see an example of deep homology in Figure 20, which illustrates the expression patterns of hox genes, specifying the body plans. On the left, the expression patterns are color-coded for C. elegans, D. melanogaster, B. floridae, M. musculus, and human. On the right, the chromosomal organization of the hox genes.
Figure 21 shows a phylogeny-based classification of the hox protein-coding genes for D. melanogaster and M. musculus with a hypothetical common ancestor. The color-coding is the same as in Figure 20.
Both Kernel and ChIN are deep homology concepts, each attempting to describe the mechanisms and categorize them.
You may also be interested in the following
Related tutorials
Pair-wise sequence alignment
Pair-wise sequence alignment methods
How to select the right substitution matrix?
References
Havstad, JC, Assis, LC, Rieppel, O. (2015). The semaphorontic view of homology. J. Exp. Zool. (Mol. Dev. Evol.) 324B: 578– 587. doi: 10.1002/jez.b.22634
IUPAC-IUB Joint Commission on Biochemical Nomenclature. Nomenclature and Symbolism for Amino Acids and Peptides: Recommendations 1983. FEBS J. 1984, 138 (1), 9–37. DOI: 10.1111/j.1432-1033.1984.tb07877.x. HTML version.
Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, Shao Y, Liechti A, Ascenção K, Rummel C, Ovchinnikova S, Mazin PV, Xenarios I, Harshman K, Mort M, Cooper DN, Sandi C, Soares MJ, Ferreira PG, Afonso S, Carneiro M, Turner JMA, VandeBerg JL, Fallahshahroudi A, Jensen P, Behr R, Lisgo S, Lindsay S, Khaitovich P, Huber W, Baker J, Anders S, Zhang YE, Kaessmann H. (2019). Gene expression across mammalian organ development. Nature. 2019 Jun 26. doi: 10.1038/s41586-019-1338-5
Descorps-Declère S, Lemoine F, Sculo Q, Lespinet O, Labedan B. (2008). The multiple facets of homology and their use in comparative genomics to study the evolution of genes, genomes, and species. Biochimie. 2008 Apr;90(4):595-608. Epub 2007 Sep 22. doi: 10.1016/j.biochi.2007.09.010
Günter P. Wagner. (2014) Homology, Genes, and Evolutionary Innovation. Princeton University Press.
Glover NM, Redestig H, Dessimoz C (2016). Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016 Jul;21(7):609-621. doi: 10.1016/j.tplants.2016.02.005. Epub 2016 Mar 22. doi: 10.1016/j.tplants.2016.02.005
Wagner, G. P. (2015). Homology and the evolutionary process: reply to Haig, Love and Brown on “Homology, Genes and Evolutionary Innovation.” Biol. & Phil. doi: 10.1007/s10539-015-9492-0
Sommer RJ. (2008). Homology and the hierarchy of biological systems. Bioessays. 2008 Jul;30(7):653-8. doi: 10.1002/bies.20776.
Tschopp P, Tabin CJ. (2017). Deep homology in the age of next-generation sequencing. Phil. Trans. R. Soc. B 372:20150475. doi: 10.1098/rstb.2015.0475.
R. Owen, On the Archetype and Homologies of the Vertebrate Skeleton, J. van Voorst, London (1847). biodiversitylibrary.org
W.M. Fitch (2001), Homology: a personal view on some of the problems, Trends Genet. 16 (2001) 227e231. doi: 10.1016/S0168-9525(00)02005-9
Walter M. Fitch (1970). Distinguishing Homologous from Analogous Proteins. Syst Zool. 1970 Jun;19(2):99-113. doi: 10.2307/2412448
Sonnhammer EL, Koonin EV (2002). Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002 Dec;18(12):619-20. doi: 10.1016/s0168-9525(02)02793-2
Gerald R Reeck, Christoph de Haen, David C Teller, Russell F Doollttle, Walter M Fitch, Richard E Dickersoq, Pierre Chambon Andrew 0 McLachlan, Emanuel argoliash Thomas H Jukes, Emile Zuckerkandl (1987). “Homology” in Proteins and Nucleic Acids: A Terminology Muddle and a Way out of It. Cell, Vol. 50, 667, August 26, 1987. doi: 10.1016/0092-8674(87)90322-9
Joseph W Thornton1, Rob DeSalle (2000). GENE FAMILY EVOLUTION AND HOMOLOGY:Genomics Meets Phylogenetics. Annu. Rev. Genomics Hum. Genet. 2000. 01:41-73. doi: 10.1146/annurev.genom.1.1.41
Peter Peneder, Barbara Wallner, Claus Vogl (2017). Exchange of genetic information between therian X and Y chromosome gametologs in old evolutionary strata. Ecol Evol. 2017 Sep 12;7(20):8478-8487. doi: 10.1002/ece3.3278. eCollection 2017 Oct. doi: 10.1002/ece3.3278
García-Moreno J, Mindell DP (2000). Rooting a phylogeny with homologous genes on opposite sex chromosomes (gametologs): a case study using avian CHD. Mol Biol Evol. 2000 Dec;17(12):1826-32. doi: 10.1093/oxfordjournals.molbev.a026283
Wolfe K (2000). Robustness--it's not where you think it is. Nat Genet. 2000 May;25(1):3-4. doi: 10.1038/75560
M F Land, R D Fernald (1992). The Evolution of Eyes. Annual Review of Neuroscience Vol. 15, 1992. doi: 10.1146/annurev.ne.15.030192.000245
Heliconius Genome Consortium (2012). Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012 Jul 5;487(7405):94-8. doi: 10.1038/nature11041
Axel Meyer (2006). Repeating Patterns of Mimicry. PLoS Biol 4(10): e341. doi: 10.1371/journal.pbio.0040341
Brower AV (2011). Hybrid speciation in Heliconius butterflies? A review and critique of the evidence. Genetica. 2011 May;139(5):589-609. 10.1007/s10709-010-9530-4
Smith J, Kronforst MR (2013). Do Heliconius butterfly species exchange mimicry alleles? Biol Lett. 2013 Jul 17;9(4):20130503. doi: 10.1098/rsbl.2013.0503
Ludwig F, Rosenthal DM, Johnston JA, Kane N, Gross BL, Lexer C, Dudley SA, Rieseberg LH, Donovan LA (2004).Selection on leaf ecophysiological traits in a desert hybrid Helianthus species and early-generation hybrids. Evolution. 2004 Dec;58(12):2682-92. doi: 10.1111/j.0014-3820.2004.tb01621.x
Jordana Neri, Tânia Wendt, Clarisse Palma-Silva (2017). Natural hybridization and genetic and morphological variation between two epiphytic bromeliads. AoB PLANTS, Volume 10, Issue 1, February 2018, plx061. doi: 10.1093/aobpla/plx061
Peter A Larsen, Steven R Hoofer, Matthew C Bozeman, Scott C Pedersen, Hugh H Genoways, Carleton J Phillips, Dorothy E Pumo, Robert J Baker (2007). Phylogenetics and Phylogeography of the Artibeus jamaicensis Complex Based on Cytochrome-b DNA Sequences. Journal of Mammalogy, Volume 88, Issue 3, June 2007, Pages 712–727. doi:L 10.1644/06-MAMM-A-125R.1
Peter A. Larsen, María R Marchán-Rivadeneira, Robert J Baker (2010). Natural hybridization generates mammalian lineage with species characteristics. Proc Natl Acad Sci U S A. 2010 Jun 22; 107(25): 11447–11452. doi: 10.1073%2Fpnas.1000133107
Schwab IR (2017). The evolution of eyes: major steps. The Keeler lecture 2017: centenary of Keeler Ltd. Eye (Lond). 2018 Feb;32(2):302-313. doi: 10.1038/eye.2017.226
Stern DL (2013). The genetic causes of convergent evolution. Nat Rev Genet. 2013 Nov;14(11):751-64. doi: 10.1038/nrg3483
Joe Parker, Georgia Tsagkogeorga, James A Cotton, Yuan Liu, Paolo Provero, Elia Stupka, Stephen J Rossiter (2013). Genome-wide signatures of convergent evolution in echolocating mammals. Nature volume 502, pages 228–231 (10 October 2013) doi: 10.1038/nature12511
Andrew D Foote, Yue Liu, Gregg W C Thomas, Tomáš Vinař, Jessica Alföldi, Jixin Deng, Shannon Dugan, Cornelis E van Elk, Margaret E Hunter, Vandita Joshi, Ziad Khan, Christie Kovar, Sandra L Lee, Kerstin Lindblad-Toh, Annalaura Mancia, Rasmus Nielsen, Xiang Qin, Jiaxin Qu, Brian J Raney, Nagarjun Vijay, Jochen B W Wolf, Matthew W Hahn, Donna M Muzny, Kim C Worley, M Thomas P Gilbert, Richard A Gibbs (2015). Convergent evolution of the genomes of marine mammals. Nature Genetics volume 47, pages 272–275 (2015) 10.1038/ng.3198
C Tristan Stayton (2015). What does convergent evolution mean? The interpretation of convergence and its implications in the search for limits to evolution. Interface Focus. 2015 Dec 6; 5(6): 20150039. doi: 10.1098/rsfs.2015.0039
Michael Y Galperin, D Roland Walker, Eugene V Koonin (1998). Analogous Enzymes: Independent Inventions in Enzyme Evolution. Genome Res. 1998. 8: 779-790. doi: 10.1101/gr.8.8.779
Mukhopadhyay B, Shukla RM, Mukhopadhyay M, Mandal KC, Haldar P, Benare A (2012). Spectrum of human tails: A report of six cases. J Indian Assoc Pediatr Surg. 2012 Jan;17(1):23-5. doi: 10.4103/0971-9261.91082
Turk CC, Kara NN, Bacanli A (2016). The Human Tail: A Simple Skin Appendage or Cutaneous Stigma of an Anomaly? Turk Neurosurg. 2016;26(1):140-5. doi: 10.5137/1019-5149.JTN.12199-14.1
Pillai MK, Nair ST (2017). A True Human Tail in a Neonate: Case report and literature review. Sultan Qaboos Univ Med J. 2017 Feb;17(1):e109-e111. doi: 10.18295/squmj.2016.17.01.020
Harris MP, Hasso SM, Ferguson MW, Fallon JF (2006). The development of archosaurian first-generation teeth in a chicken mutant. Curr Biol. 2006 Feb 21;16(4):371-7. doi: 10.1016/j.cub.2005.12.047
Butler AB, Saidel WM (2000). Defining sameness: historical, biological, and generative homology. Bioessays. 2000 Sep;22(9):846-53. doi: 10.1002/1521-1878...
Arendt D (2008). The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet. 2008 Nov;9(11):868-82. doi: 10.1038/nrg2416. doi: 10.1038/nrg2416
Davidson EH, Erwin DH (2006). Gene regulatory networks and the evolution of animal body plans. Science. 2006 Feb 10;311(5762):796-800. doi: 10.1126/science.1113832
Owen R (1843). Lectures on the comparative anatomy and physiology of the invertebrate animals, delivered at the Royal College of Surgeons in 1843. London, UK: Longman, Brown, Green & Longmans. doi: 10.5962/bhl.title.11742
Davidson EH, Erwin DH (2006). Gene regulatory networks and the evolution of animal body plans. Science. 2006 Feb 10;311(5762):796-800. doi: 10.1126/science.1113832
Bruneau BG (2013). Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013 Mar 1;5(3):a008292. doi: 10.1101/cshperspect.a008292
Assis R, Zhou Q, Bachtrog D (2012). Sex-biased transcriptome evolution in Drosophila. Genome Biol Evol. 2012;4(11):1189-200. doi: 10.1093/gbe/evs093
Romero IG, Ruvinsky I, Gilad Y (2012). Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012 Jun 18;13(7):505-16. doi: 10.1038/nrg3229
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013 Aug;8(8):1494-512. doi: 10.1038/nprot.2013.084
Sheaffer KL, Kaestner KH (2012). Transcriptional networks in liver and intestinal development. Cold Spring Harb Perspect Biol. 2012 Sep 1;4(9):a008284. doi: 10.1101/cshperspect.a008284
Shubin N, Tabin C, Carroll S (1997). Fossils, genes and the evolution of animal limbs. Nature. 1997 Aug 14;388(6643):639-48. 10.1038/41710
Hueber SD, Weiller GF, Djordjevic MA, Frickey T (2010) Improving Hox Protein Classification across the Major Model Organisms. PLoS ONE 5(5): e10820. doi: 10.1371/journal.pone.0010820